Jichao Zhao
Dynamic Position Transformation and Boundary Refinement Network for Left Atrial Segmentation
Jul 07, 2024Abstract:Left atrial (LA) segmentation is a crucial technique for irregular heartbeat (i.e., atrial fibrillation) diagnosis. Most current methods for LA segmentation strictly assume that the input data is acquired using object-oriented center cropping, while this assumption may not always hold in practice due to the high cost of manual object annotation. Random cropping is a straightforward data pre-processing approach. However, it 1) introduces significant irregularities and incompleteness in the input data and 2) disrupts the coherence and continuity of object boundary regions. To tackle these issues, we propose a novel Dynamic Position transformation and Boundary refinement Network (DPBNet). The core idea is to dynamically adjust the relative position of irregular targets to construct their contextual relationships and prioritize difficult boundary pixels to enhance foreground-background distinction. Specifically, we design a shuffle-then-reorder attention module to adjust the position of disrupted objects in the latent space using dynamic generation ratios, such that the vital dependencies among these random cropping targets could be well captured and preserved. Moreover, to improve the accuracy of boundary localization, we introduce a dual fine-grained boundary loss with scenario-adaptive weights to handle the ambiguity of the dual boundary at a fine-grained level, promoting the clarity and continuity of the obtained results. Extensive experimental results on benchmark dataset have demonstrated that DPBNet consistently outperforms existing state-of-the-art methods.
A Global Benchmark of Algorithms for Segmenting Late Gadolinium-Enhanced Cardiac Magnetic Resonance Imaging
May 07, 2020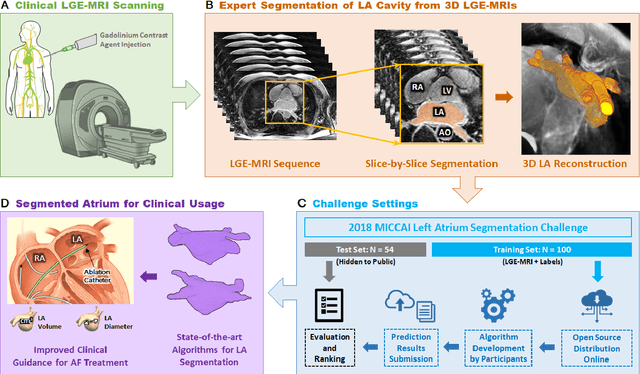
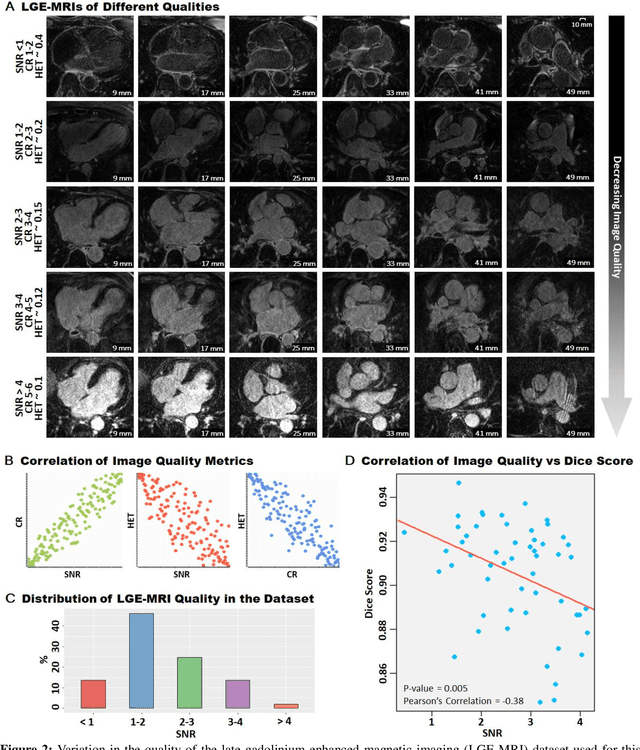
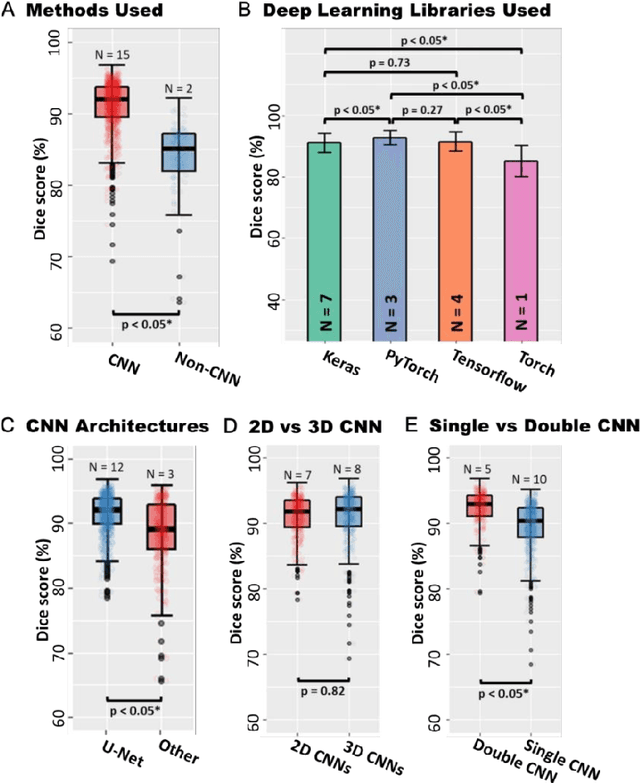
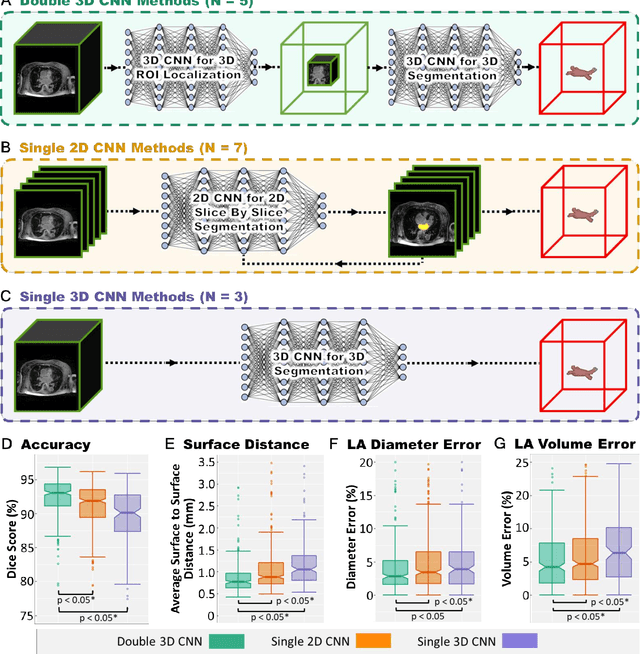
Abstract:Segmentation of cardiac images, particularly late gadolinium-enhanced magnetic resonance imaging (LGE-MRI) widely used for visualizing diseased cardiac structures, is a crucial first step for clinical diagnosis and treatment. However, direct segmentation of LGE-MRIs is challenging due to its attenuated contrast. Since most clinical studies have relied on manual and labor-intensive approaches, automatic methods are of high interest, particularly optimized machine learning approaches. To address this, we organized the "2018 Left Atrium Segmentation Challenge" using 154 3D LGE-MRIs, currently the world's largest cardiac LGE-MRI dataset, and associated labels of the left atrium segmented by three medical experts, ultimately attracting the participation of 27 international teams. In this paper, extensive analysis of the submitted algorithms using technical and biological metrics was performed by undergoing subgroup analysis and conducting hyper-parameter analysis, offering an overall picture of the major design choices of convolutional neural networks (CNNs) and practical considerations for achieving state-of-the-art left atrium segmentation. Results show the top method achieved a dice score of 93.2% and a mean surface to a surface distance of 0.7 mm, significantly outperforming prior state-of-the-art. Particularly, our analysis demonstrated that double, sequentially used CNNs, in which a first CNN is used for automatic region-of-interest localization and a subsequent CNN is used for refined regional segmentation, achieved far superior results than traditional methods and pipelines containing single CNNs. This large-scale benchmarking study makes a significant step towards much-improved segmentation methods for cardiac LGE-MRIs, and will serve as an important benchmark for evaluating and comparing the future works in the field.
Segmentation of histological images and fibrosis identification with a convolutional neural network
Mar 20, 2018



Abstract:Segmentation of histological images is one of the most crucial tasks for many biomedical analyses including quantification of certain tissue type. However, challenges are posed by high variability and complexity of structural features in such images, in addition to imaging artifacts. Further, the conventional approach of manual thresholding is labor-intensive, and highly sensitive to inter- and intra-image intensity variations. An accurate and robust automated segmentation method is of high interest. We propose and evaluate an elegant convolutional neural network (CNN) designed for segmentation of histological images, particularly those with Masson's trichrome stain. The network comprises of 11 successive convolutional - rectified linear unit - batch normalization layers, and outperformed state-of-the-art CNNs on a dataset of cardiac histological images (labeling fibrosis, myocytes, and background) with a Dice similarity coefficient of 0.947. With 100 times fewer (only 300 thousand) trainable parameters, our CNN is less susceptible to overfitting, and is efficient. Additionally, it retains image resolution from input to output, captures fine-grained details, and can be trained end-to-end smoothly. To the best of our knowledge, this is the first deep CNN tailored for the problem of concern, and may be extended to solve similar segmentation tasks to facilitate investigations into pathology and clinical treatment.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge