Iuri Macocco
Outlier dimensions favor frequent tokens in language model
Mar 27, 2025Abstract:We study last-layer outlier dimensions, i.e.dimensions that display extreme activations for the majority of inputs. We show that outlier dimensions arise in many different modern language models, and trace their function back to the heuristic of constantly predicting frequent words. We further show how a model can block this heuristic when it is not contextually appropriate, by assigning a counterbalancing weight mass to the remaining dimensions, and we investigate which model parameters boost outlier dimensions and when they arise during training. We conclude that outlier dimensions are a specialized mechanism discovered by many distinct models to implement a useful token prediction heuristic.
Beyond the noise: intrinsic dimension estimation with optimal neighbourhood identification
May 24, 2024



Abstract:The Intrinsic Dimension (ID) is a key concept in unsupervised learning and feature selection, as it is a lower bound to the number of variables which are necessary to describe a system. However, in almost any real-world dataset the ID depends on the scale at which the data are analysed. Quite typically at a small scale, the ID is very large, as the data are affected by measurement errors. At large scale, the ID can also be erroneously large, due to the curvature and the topology of the manifold containing the data. In this work, we introduce an automatic protocol to select the sweet spot, namely the correct range of scales in which the ID is meaningful and useful. This protocol is based on imposing that for distances smaller than the correct scale the density of the data is constant. Since to estimate the density it is necessary to know the ID, this condition is imposed self-consistently. We illustrate the usefulness and robustness of this procedure by benchmarks on artificial and real-world datasets.
Emergence of a High-Dimensional Abstraction Phase in Language Transformers
May 24, 2024Abstract:A language model (LM) is a mapping from a linguistic context to an output token. However, much remains to be known about this mapping, including how its geometric properties relate to its function. We take a high-level geometric approach to its analysis, observing, across five pre-trained transformer-based LMs and three input datasets, a distinct phase characterized by high intrinsic dimensionality. During this phase, representations (1) correspond to the first full linguistic abstraction of the input; (2) are the first to viably transfer to downstream tasks; (3) predict each other across different LMs. Moreover, we find that an earlier onset of the phase strongly predicts better language modelling performance. In short, our results suggest that a central high-dimensionality phase underlies core linguistic processing in many common LM architectures.
Intrinsic dimension estimation for discrete metrics
Jul 20, 2022



Abstract:Real world-datasets characterized by discrete features are ubiquitous: from categorical surveys to clinical questionnaires, from unweighted networks to DNA sequences. Nevertheless, the most common unsupervised dimensional reduction methods are designed for continuous spaces, and their use for discrete spaces can lead to errors and biases. In this letter we introduce an algorithm to infer the intrinsic dimension (ID) of datasets embedded in discrete spaces. We demonstrate its accuracy on benchmark datasets, and we apply it to analyze a metagenomic dataset for species fingerprinting, finding a surprisingly small ID, of order 2. This suggests that evolutive pressure acts on a low-dimensional manifold despite the high-dimensionality of sequences' space.
DADApy: Distance-based Analysis of DAta-manifolds in Python
May 04, 2022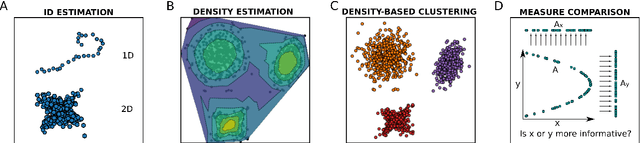
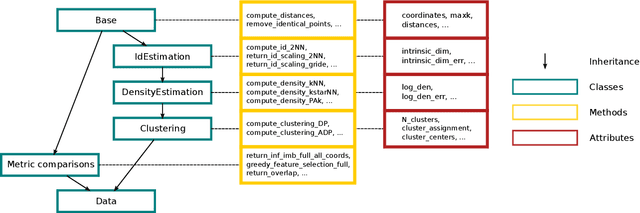
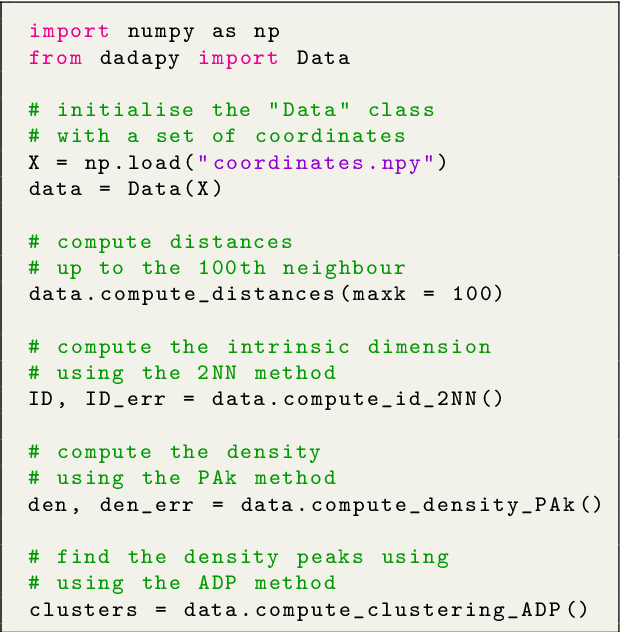
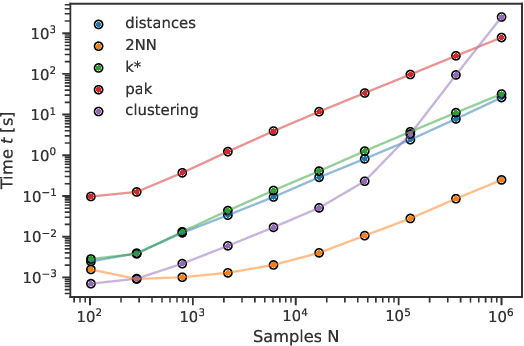
Abstract:DADApy is a python software package for analysing and characterising high-dimensional data manifolds. It provides methods for estimating the intrinsic dimension and the probability density, for performing density-based clustering and for comparing different distance metrics. We review the main functionalities of the package and exemplify its usage in toy cases and in a real-world application. The package is freely available under the open-source Apache 2.0 license and can be downloaded from the Github page https://github.com/sissa-data-science/DADApy.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge