Hongxin Ding
3DS: Decomposed Difficulty Data Selection's Case Study on LLM Medical Domain Adaptation
Oct 13, 2024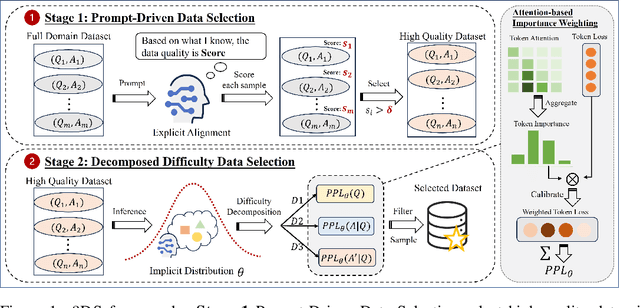
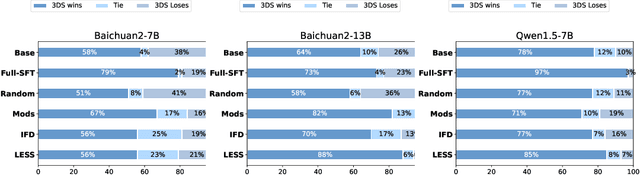
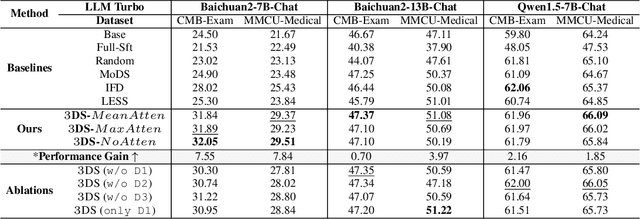
Abstract:Large Language Models(LLMs) excel in general tasks but struggle in specialized domains like healthcare due to limited domain-specific knowledge.Supervised Fine-Tuning(SFT) data construction for domain adaptation often relies on heuristic methods, such as GPT-4 annotation or manual data selection, with a data-centric focus on presumed diverse, high-quality datasets. However, these methods overlook the model's inherent knowledge distribution, introducing noise, redundancy, and irrelevant data, leading to a mismatch between the selected data and the model's learning task, resulting in suboptimal performance. To address this, we propose a two-stage model-centric data selection framework, Decomposed Difficulty Data Selection (3DS), which aligns data with the model's knowledge distribution for optimized adaptation. In Stage1, we apply Prompt-Driven Data Selection via Explicit Alignment, where the the model filters irrelevant or redundant data based on its internal knowledge. In Stage2, we perform Decomposed Difficulty Data Selection, where data selection is guided by our defined difficulty decomposition, using three metrics: Instruction Understanding, Response Confidence, and Response Correctness. Additionally, an attention-based importance weighting mechanism captures token importance for more accurate difficulty calibration. This two-stage approach ensures the selected data is not only aligned with the model's knowledge and preferences but also appropriately challenging for the model to learn, leading to more effective and targeted domain adaptation. In the case study of the medical domain, our extensive experiments on real-world healthcare datasets demonstrate the superiority of 3DS over exisiting methods in accuracy by over 5.29%. Our dataset and code will be open-sourced at https://anonymous.4open.science/r/3DS-E67F.
Think and Retrieval: A Hypothesis Knowledge Graph Enhanced Medical Large Language Models
Dec 26, 2023Abstract:We explore how the rise of Large Language Models (LLMs) significantly impacts task performance in the field of Natural Language Processing. We focus on two strategies, Retrieval-Augmented Generation (RAG) and Fine-Tuning (FT), and propose the Hypothesis Knowledge Graph Enhanced (HyKGE) framework, leveraging a knowledge graph to enhance medical LLMs. By integrating LLMs and knowledge graphs, HyKGE demonstrates superior performance in addressing accuracy and interpretability challenges, presenting potential applications in the medical domain. Our evaluations using real-world datasets highlight HyKGE's superiority in providing accurate knowledge with precise confidence, particularly in complex and difficult scenarios. The code will be available until published.
A ModelOps-based Framework for Intelligent Medical Knowledge Extraction
Oct 04, 2023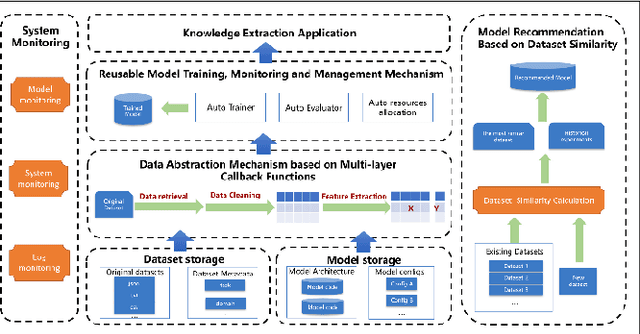
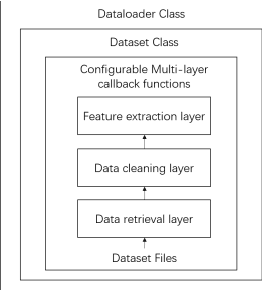
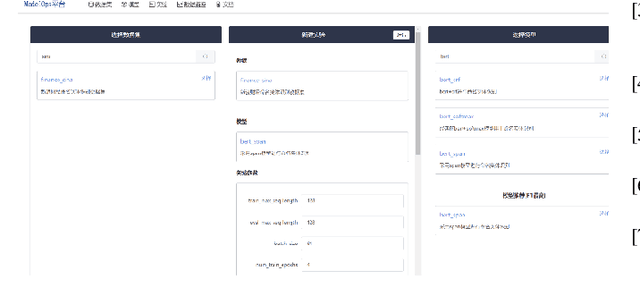
Abstract:Extracting medical knowledge from healthcare texts enhances downstream tasks like medical knowledge graph construction and clinical decision-making. However, the construction and application of knowledge extraction models lack automation, reusability and unified management, leading to inefficiencies for researchers and high barriers for non-AI experts such as doctors, to utilize knowledge extraction. To address these issues, we propose a ModelOps-based intelligent medical knowledge extraction framework that offers a low-code system for model selection, training, evaluation and optimization. Specifically, the framework includes a dataset abstraction mechanism based on multi-layer callback functions, a reusable model training, monitoring and management mechanism. We also propose a model recommendation method based on dataset similarity, which helps users quickly find potentially suitable models for a given dataset. Our framework provides convenience for researchers to develop models and simplifies model access for non-AI experts such as doctors.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge