Haanju Yoo
HyperCLOVA X Technical Report
Apr 13, 2024Abstract:We introduce HyperCLOVA X, a family of large language models (LLMs) tailored to the Korean language and culture, along with competitive capabilities in English, math, and coding. HyperCLOVA X was trained on a balanced mix of Korean, English, and code data, followed by instruction-tuning with high-quality human-annotated datasets while abiding by strict safety guidelines reflecting our commitment to responsible AI. The model is evaluated across various benchmarks, including comprehensive reasoning, knowledge, commonsense, factuality, coding, math, chatting, instruction-following, and harmlessness, in both Korean and English. HyperCLOVA X exhibits strong reasoning capabilities in Korean backed by a deep understanding of the language and cultural nuances. Further analysis of the inherent bilingual nature and its extension to multilingualism highlights the model's cross-lingual proficiency and strong generalization ability to untargeted languages, including machine translation between several language pairs and cross-lingual inference tasks. We believe that HyperCLOVA X can provide helpful guidance for regions or countries in developing their sovereign LLMs.
Development of deep biological ages aware of morbidity and mortality based on unsupervised and semi-supervised deep learning approaches
Feb 01, 2023



Abstract:Background: While deep learning technology, which has the capability of obtaining latent representations based on large-scale data, can be a potential solution for the discovery of a novel aging biomarker, existing deep learning methods for biological age estimation usually depend on chronological ages and lack of consideration of mortality and morbidity that are the most significant outcomes of aging. Methods: This paper proposes a novel deep learning model to learn latent representations of biological aging in regard to subjects' morbidity and mortality. The model utilizes health check-up data in addition to morbidity and mortality information to learn the complex relationships between aging and measured clinical attributes. Findings: The proposed model is evaluated on a large dataset of general populations compared with KDM and other learning-based models. Results demonstrate that biological ages obtained by the proposed model have superior discriminability of subjects' morbidity and mortality.
MED-SE: Medical Entity Definition-based Sentence Embedding
Dec 09, 2022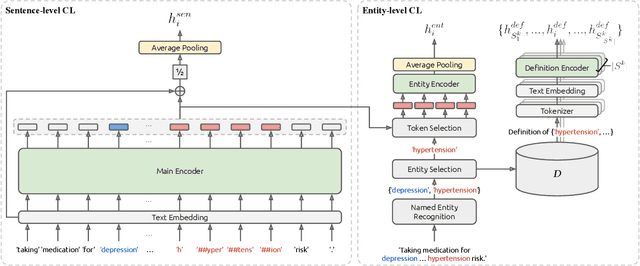
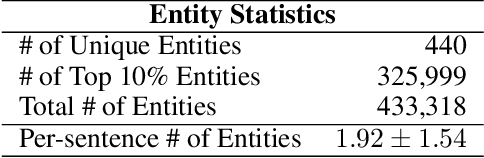
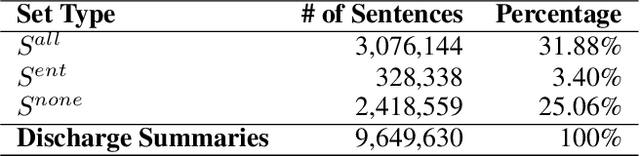
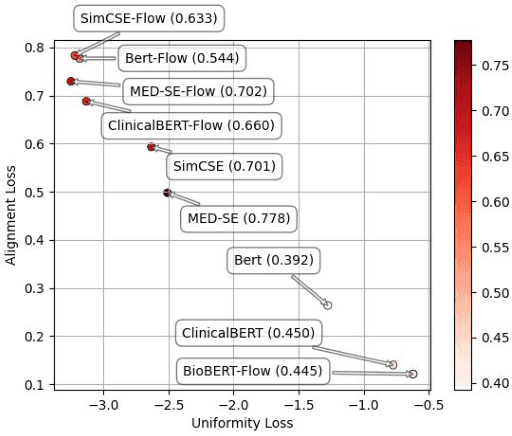
Abstract:We propose Medical Entity Definition-based Sentence Embedding (MED-SE), a novel unsupervised contrastive learning framework designed for clinical texts, which exploits the definitions of medical entities. To this end, we conduct an extensive analysis of multiple sentence embedding techniques in clinical semantic textual similarity (STS) settings. In the entity-centric setting that we have designed, MED-SE achieves significantly better performance, while the existing unsupervised methods including SimCSE show degraded performance. Our experiments elucidate the inherent discrepancies between the general- and clinical-domain texts, and suggest that entity-centric contrastive approaches may help bridge this gap and lead to a better representation of clinical sentences.
An Automatic ICD Coding Network Using Partition-Based Label Attention
Nov 15, 2022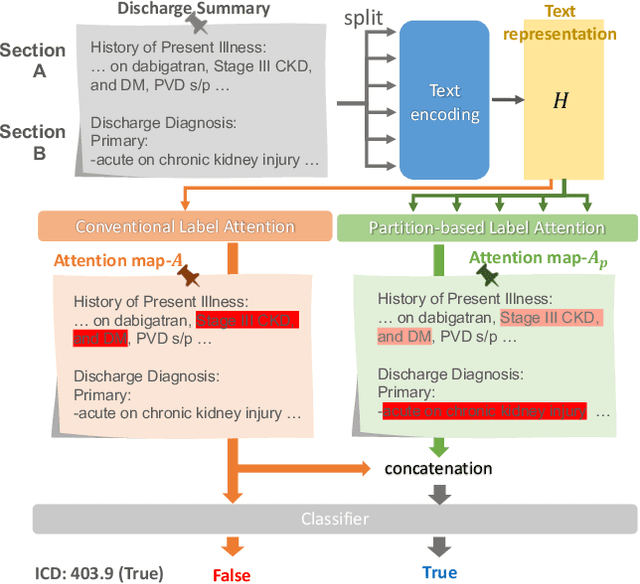
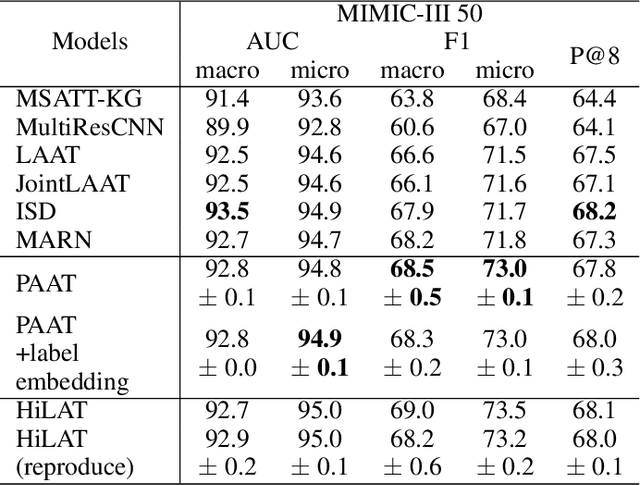
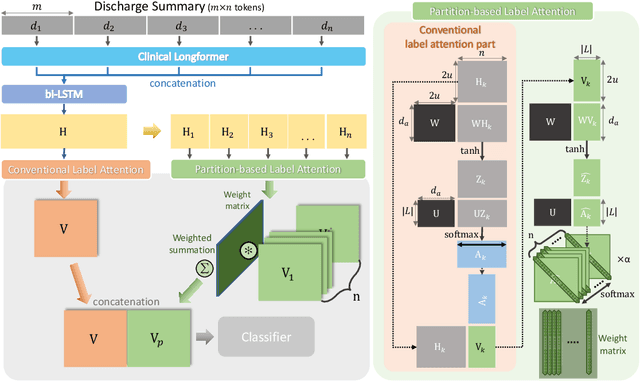
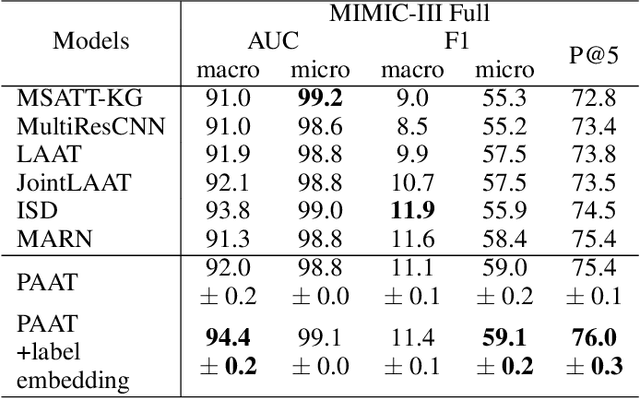
Abstract:International Classification of Diseases (ICD) is a global medical classification system which provides unique codes for diagnoses and procedures appropriate to a patient's clinical record. However, manual coding by human coders is expensive and error-prone. Automatic ICD coding has the potential to solve this problem. With the advancement of deep learning technologies, many deep learning-based methods for automatic ICD coding are being developed. In particular, a label attention mechanism is effective for multi-label classification, i.e., the ICD coding. It effectively obtains the label-specific representations from the input clinical records. However, because the existing label attention mechanism finds key tokens in the entire text at once, the important information dispersed in each paragraph may be omitted from the attention map. To overcome this, we propose a novel neural network architecture composed of two parts of encoders and two kinds of label attention layers. The input text is segmentally encoded in the former encoder and integrated by the follower. Then, the conventional and partition-based label attention mechanisms extract important global and local feature representations. Our classifier effectively integrates them to enhance the ICD coding performance. We verified the proposed method using the MIMIC-III, a benchmark dataset of the ICD coding. Our results show that our network improves the ICD coding performance based on the partition-based mechanism.
Cross-modal Variational Auto-encoder with Distributed Latent Spaces and Associators
May 30, 2019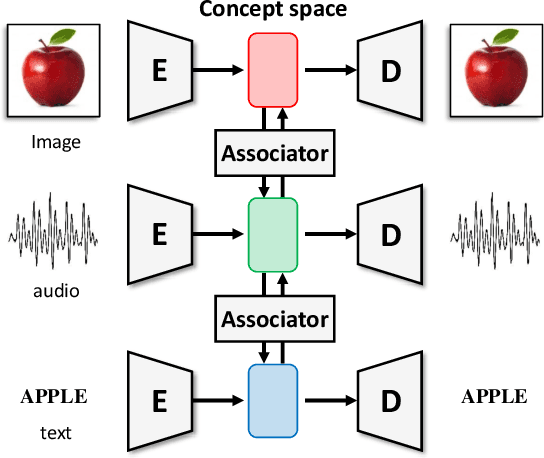
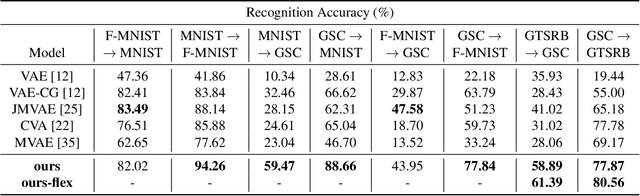
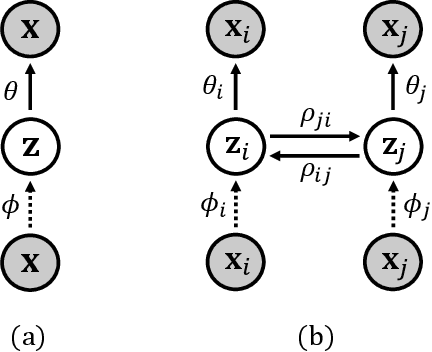
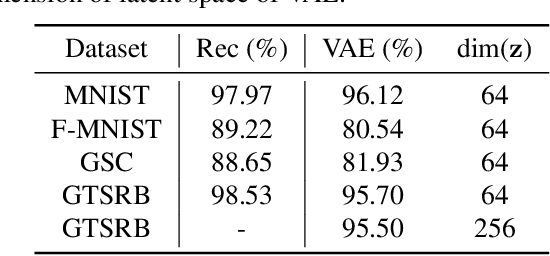
Abstract:In this paper, we propose a novel structure for a cross-modal data association, which is inspired by the recent research on the associative learning structure of the brain. We formulate the cross-modal association in Bayesian inference framework realized by a deep neural network with multiple variational auto-encoders and variational associators. The variational associators transfer the latent spaces between auto-encoders that represent different modalities. The proposed structure successfully associates even heterogeneous modal data and easily incorporates the additional modality to the entire network via the proposed cross-modal associator. Furthermore, the proposed structure can be trained with only a small amount of paired data since auto-encoders can be trained by unsupervised manner. Through experiments, the effectiveness of the proposed structure is validated on various datasets including visual and auditory data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge