Ha Q. Nguyen
Evaluating the impact of an explainable machine learning system on the interobserver agreement in chest radiograph interpretation
Apr 01, 2023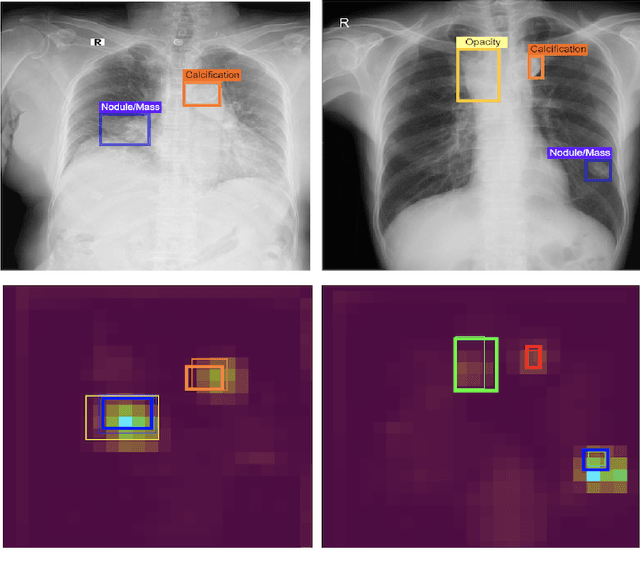
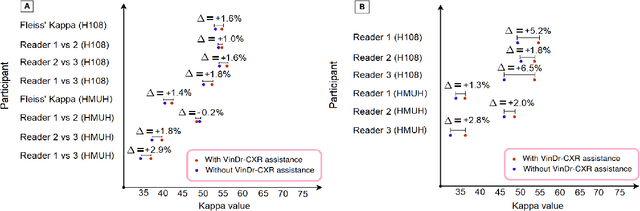
Abstract:We conducted a prospective study to measure the clinical impact of an explainable machine learning system on interobserver agreement in chest radiograph interpretation. The AI system, which we call as it VinDr-CXR when used as a diagnosis-supporting tool, significantly improved the agreement between six radiologists with an increase of 1.5% in mean Fleiss' Kappa. In addition, we also observed that, after the radiologists consulted AI's suggestions, the agreement between each radiologist and the system was remarkably increased by 3.3% in mean Cohen's Kappa. This work has been accepted for publication in IEEE Access and this paper is our short version submitted to the Midwest Machine Learning Symposium (MMLS 2023), Chicago, IL, USA.
Improving Object Detection in Medical Image Analysis through Multiple Expert Annotators: An Empirical Investigation
Mar 29, 2023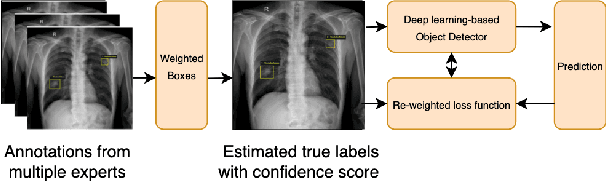

Abstract:The work discusses the use of machine learning algorithms for anomaly detection in medical image analysis and how the performance of these algorithms depends on the number of annotators and the quality of labels. To address the issue of subjectivity in labeling with a single annotator, we introduce a simple and effective approach that aggregates annotations from multiple annotators with varying levels of expertise. We then aim to improve the efficiency of predictive models in abnormal detection tasks by estimating hidden labels from multiple annotations and using a re-weighted loss function to improve detection performance. Our method is evaluated on a real-world medical imaging dataset and outperforms relevant baselines that do not consider disagreements among annotators.
Learning to diagnose common thorax diseases on chest radiographs from radiology reports in Vietnamese
Sep 11, 2022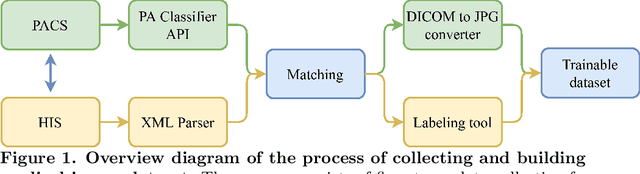
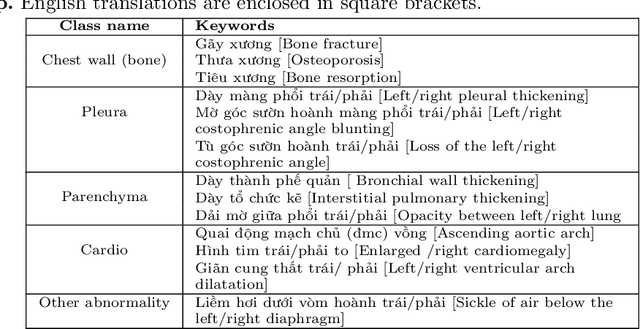
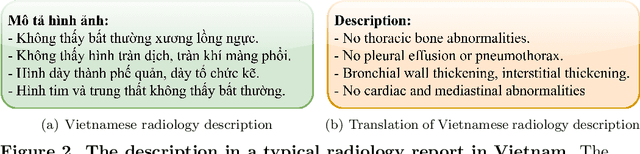
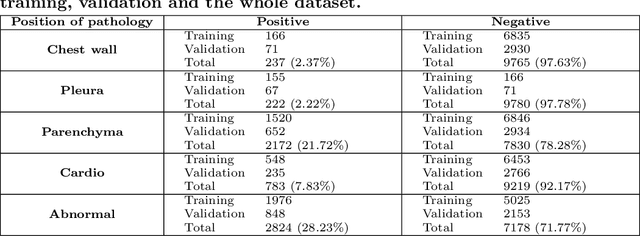
Abstract:We propose a data collecting and annotation pipeline that extracts information from Vietnamese radiology reports to provide accurate labels for chest X-ray (CXR) images. This can benefit Vietnamese radiologists and clinicians by annotating data that closely match their endemic diagnosis categories which may vary from country to country. To assess the efficacy of the proposed labeling technique, we built a CXR dataset containing 9,752 studies and evaluated our pipeline using a subset of this dataset. With an F1-score of at least 0.9923, the evaluation demonstrates that our labeling tool performs precisely and consistently across all classes. After building the dataset, we train deep learning models that leverage knowledge transferred from large public CXR datasets. We employ a variety of loss functions to overcome the curse of imbalanced multi-label datasets and conduct experiments with various model architectures to select the one that delivers the best performance. Our best model (CheXpert-pretrained EfficientNet-B2) yields an F1-score of 0.6989 (95% CI 0.6740, 0.7240), AUC of 0.7912, sensitivity of 0.7064 and specificity of 0.8760 for the abnormal diagnosis in general. Finally, we demonstrate that our coarse classification (based on five specific locations of abnormalities) yields comparable results to fine classification (twelve pathologies) on the benchmark CheXpert dataset for general anomaly detection while delivering better performance in terms of the average performance of all classes.
An Accurate and Explainable Deep Learning System Improves Interobserver Agreement in the Interpretation of Chest Radiograph
Aug 06, 2022Abstract:Recent artificial intelligence (AI) algorithms have achieved radiologist-level performance on various medical classification tasks. However, only a few studies addressed the localization of abnormal findings from CXR scans, which is essential in explaining the image-level classification to radiologists. We introduce in this paper an explainable deep learning system called VinDr-CXR that can classify a CXR scan into multiple thoracic diseases and, at the same time, localize most types of critical findings on the image. VinDr-CXR was trained on 51,485 CXR scans with radiologist-provided bounding box annotations. It demonstrated a comparable performance to experienced radiologists in classifying 6 common thoracic diseases on a retrospective validation set of 3,000 CXR scans, with a mean area under the receiver operating characteristic curve (AUROC) of 0.967 (95% confidence interval [CI]: 0.958-0.975). The VinDr-CXR was also externally validated in independent patient cohorts and showed its robustness. For the localization task with 14 types of lesions, our free-response receiver operating characteristic (FROC) analysis showed that the VinDr-CXR achieved a sensitivity of 80.2% at the rate of 1.0 false-positive lesion identified per scan. A prospective study was also conducted to measure the clinical impact of the VinDr-CXR in assisting six experienced radiologists. The results indicated that the proposed system, when used as a diagnosis supporting tool, significantly improved the agreement between radiologists themselves with an increase of 1.5% in mean Fleiss' Kappa. We also observed that, after the radiologists consulted VinDr-CXR's suggestions, the agreement between each of them and the system was remarkably increased by 3.3% in mean Cohen's Kappa.
Slice-level Detection of Intracranial Hemorrhage on CT Using Deep Descriptors of Adjacent Slices
Aug 05, 2022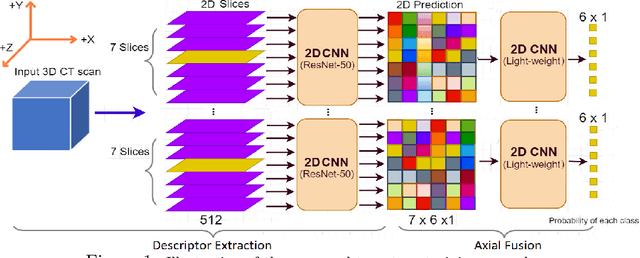
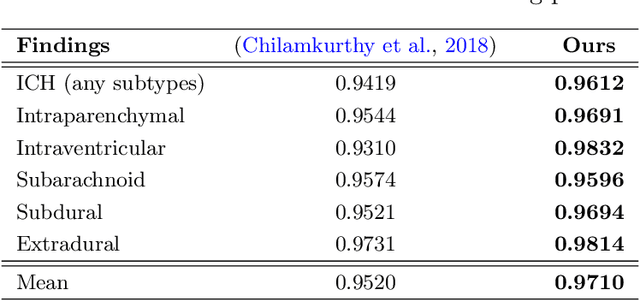
Abstract:The rapid development in representation learning techniques and the availability of large-scale medical imaging data have to a rapid increase in the use of machine learning in the 3D medical image analysis. In particular, deep convolutional neural networks (D-CNNs) have been key players and were adopted by the medical imaging community to assist clinicians and medical experts in disease diagnosis. However, training deep neural networks such as D-CNN on high-resolution 3D volumes of Computed Tomography (CT) scans for diagnostic tasks poses formidable computational challenges. This raises the need of developing deep learning-based approaches that are robust in learning representations in 2D images, instead 3D scans. In this paper, we propose a new strategy to train \emph{slice-level} classifiers on CT scans based on the descriptors of the adjacent slices along the axis. In particular, each of which is extracted through a convolutional neural network (CNN). This method is applicable to CT datasets with per-slice labels such as the RSNA Intracranial Hemorrhage (ICH) dataset, which aims to predict the presence of ICH and classify it into 5 different sub-types. We obtain a single model in the top 4\% best-performing solutions of the RSNA ICH challenge, where model ensembles are allowed. Experiments also show that the proposed method significantly outperforms the baseline model on CQ500. The proposed method is general and can be applied for other 3D medical diagnosis tasks such as MRI imaging. To encourage new advances in the field, we will make our codes and pre-trained model available upon acceptance of the paper.
Phase Recognition in Contrast-Enhanced CT Scans based on Deep Learning and Random Sampling
Mar 20, 2022Abstract:A fully automated system for interpreting abdominal computed tomography (CT) scans with multiple phases of contrast enhancement requires an accurate classification of the phases. This work aims at developing and validating a precise, fast multi-phase classifier to recognize three main types of contrast phases in abdominal CT scans. We propose in this study a novel method that uses a random sampling mechanism on top of deep CNNs for the phase recognition of abdominal CT scans of four different phases: non-contrast, arterial, venous, and others. The CNNs work as a slice-wise phase prediction, while the random sampling selects input slices for the CNN models. Afterward, majority voting synthesizes the slice-wise results of the CNNs, to provide the final prediction at scan level. Our classifier was trained on 271,426 slices from 830 phase-annotated CT scans, and when combined with majority voting on 30% of slices randomly chosen from each scan, achieved a mean F1-score of 92.09% on our internal test set of 358 scans. The proposed method was also evaluated on 2 external test sets: CTPAC-CCRCC (N = 242) and LiTS (N = 131), which were annotated by our experts. Although a drop in performance has been observed, the model performance remained at a high level of accuracy with a mean F1-score of 76.79% and 86.94% on CTPAC-CCRCC and LiTS datasets, respectively. Our experimental results also showed that the proposed method significantly outperformed the state-of-the-art 3D approaches while requiring less computation time for inference.
VinDr-Mammo: A large-scale benchmark dataset for computer-aided diagnosis in full-field digital mammography
Mar 20, 2022Abstract:Mammography, or breast X-ray, is the most widely used imaging modality to detect cancer and other breast diseases. Recent studies have shown that deep learning-based computer-assisted detection and diagnosis (CADe or CADx) tools have been developed to support physicians and improve the accuracy of interpreting mammography. However, most published datasets of mammography are either limited on sample size or digitalized from screen-film mammography (SFM), hindering the development of CADe and CADx tools which are developed based on full-field digital mammography (FFDM). To overcome this challenge, we introduce VinDr-Mammo - a new benchmark dataset of FFDM for detecting and diagnosing breast cancer and other diseases in mammography. The dataset consists of 5,000 mammography exams, each of which has four standard views and is double read with disagreement (if any) being resolved by arbitration. It is created for the assessment of Breast Imaging Reporting and Data System (BI-RADS) and density at the breast level. In addition, the dataset also provides the category, location, and BI-RADS assessment of non-benign findings. We make VinDr-Mammo publicly available on PhysioNet as a new imaging resource to promote advances in developing CADe and CADx tools for breast cancer screening.
VinDr-PCXR: An open, large-scale chest radiograph dataset for interpretation of common thoracic diseases in children
Mar 20, 2022
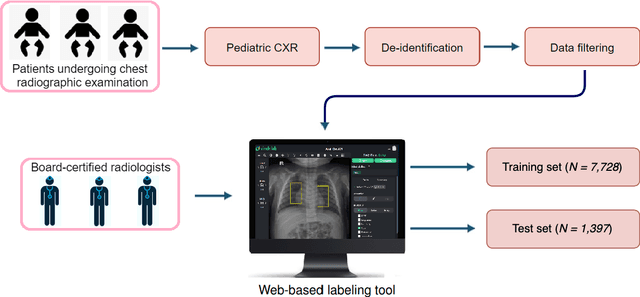
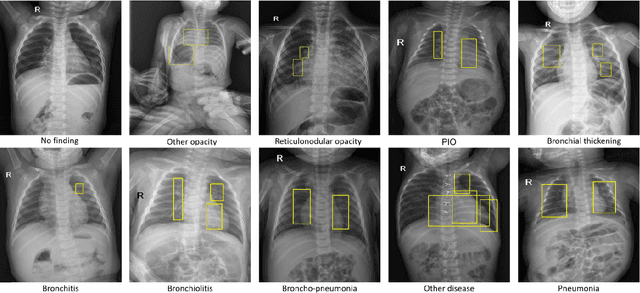
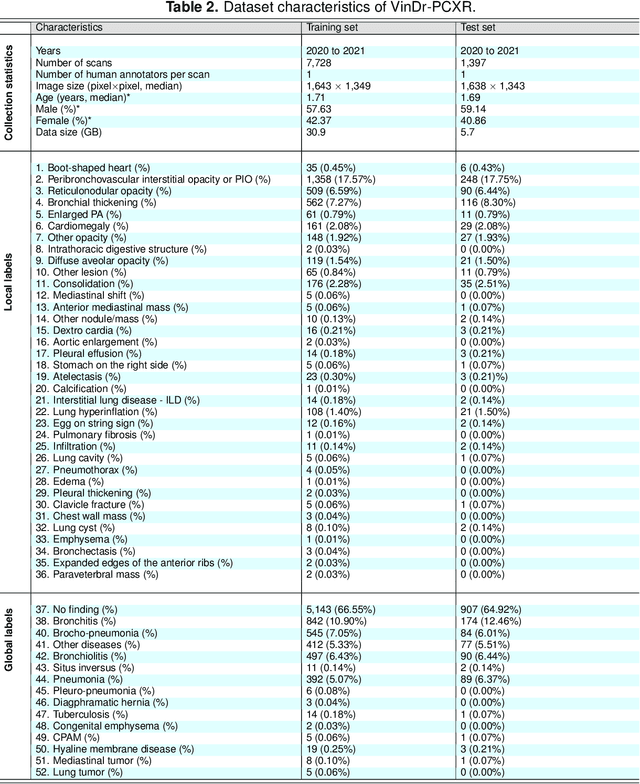
Abstract:Computer-aided diagnosis systems in adult chest radiography (CXR) have recently achieved great success thanks to the availability of large-scale, annotated datasets and the advent of high-performance supervised learning algorithms. However, the development of diagnostic models for detecting and diagnosing pediatric diseases in CXR scans is undertaken due to the lack of high-quality physician-annotated datasets. To overcome this challenge, we introduce and release VinDr-PCXR, a new pediatric CXR dataset of 9,125 studies retrospectively collected from a major pediatric hospital in Vietnam between 2020 and 2021. Each scan was manually annotated by a pediatric radiologist who has more than ten years of experience. The dataset was labeled for the presence of 36 critical findings and 15 diseases. In particular, each abnormal finding was identified via a rectangle bounding box on the image. To the best of our knowledge, this is the first and largest pediatric CXR dataset containing lesion-level annotations and image-level labels for the detection of multiple findings and diseases. For algorithm development, the dataset was divided into a training set of 7,728 and a test set of 1,397. To encourage new advances in pediatric CXR interpretation using data-driven approaches, we provide a detailed description of the VinDr-PCXR data sample and make the dataset publicly available on https://physionet.org/.
Learning from Multiple Expert Annotators for Enhancing Anomaly Detection in Medical Image Analysis
Mar 20, 2022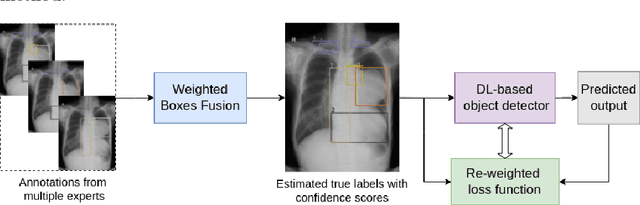
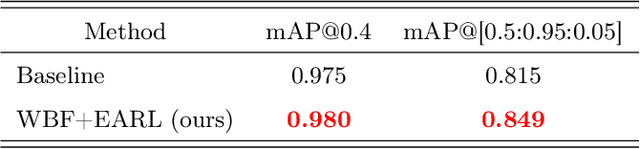
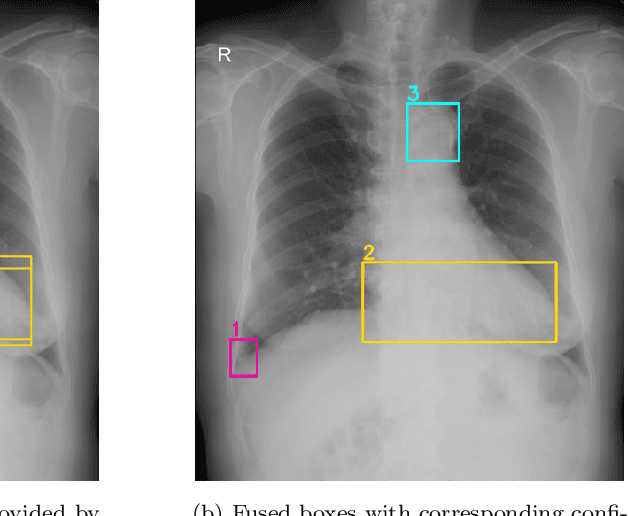
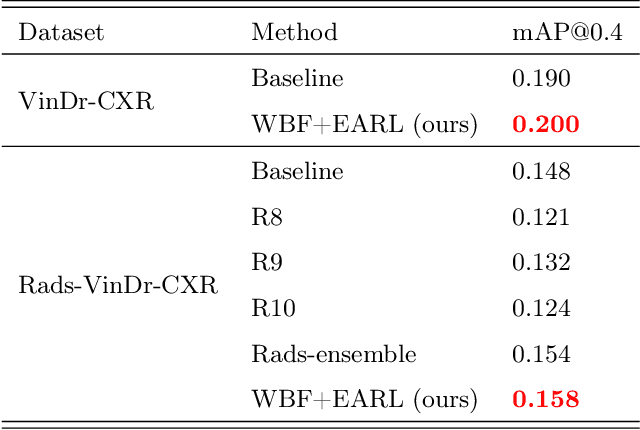
Abstract:Building an accurate computer-aided diagnosis system based on data-driven approaches requires a large amount of high-quality labeled data. In medical imaging analysis, multiple expert annotators often produce subjective estimates about "ground truth labels" during the annotation process, depending on their expertise and experience. As a result, the labeled data may contain a variety of human biases with a high rate of disagreement among annotators, which significantly affect the performance of supervised machine learning algorithms. To tackle this challenge, we propose a simple yet effective approach to combine annotations from multiple radiology experts for training a deep learning-based detector that aims to detect abnormalities on medical scans. The proposed method first estimates the ground truth annotations and confidence scores of training examples. The estimated annotations and their scores are then used to train a deep learning detector with a re-weighted loss function to localize abnormal findings. We conduct an extensive experimental evaluation of the proposed approach on both simulated and real-world medical imaging datasets. The experimental results show that our approach significantly outperforms baseline approaches that do not consider the disagreements among annotators, including methods in which all of the noisy annotations are treated equally as ground truth and the ensemble of different models trained on different label sets provided separately by annotators.
Transparency strategy-based data augmentation for BI-RADS classification of mammograms
Mar 20, 2022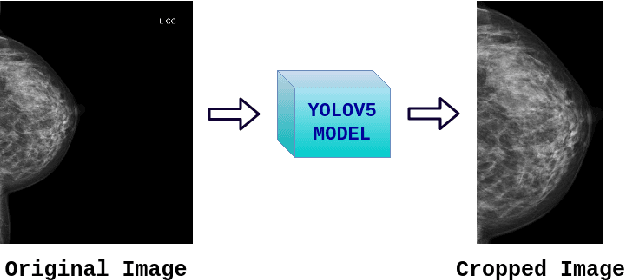
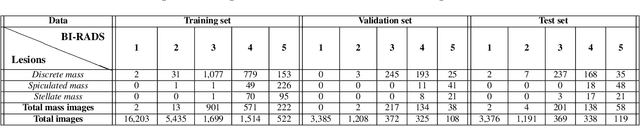
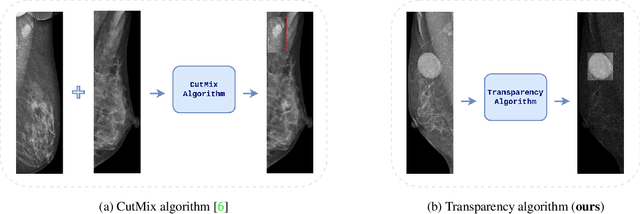
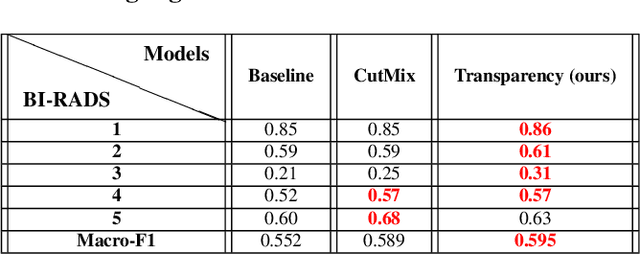
Abstract:Image augmentation techniques have been widely investigated to improve the performance of deep learning (DL) algorithms on mammography classification tasks. Recent methods have proved the efficiency of image augmentation on data deficiency or data imbalance issues. In this paper, we propose a novel transparency strategy to boost the Breast Imaging Reporting and Data System (BI-RADS) scores of mammograms classifier. The proposed approach utilizes the Region of Interest (ROI) information to generate more high-risk training examples from original images. Our extensive experiments were conducted on our benchmark mammography dataset. The experiment results show that the proposed approach surpasses current state-of-the-art data augmentation techniques such as Upsampling or CutMix. The study highlights that the transparency method is more effective than other augmentation strategies for BI-RADS classification and can be widely applied for our computer vision tasks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge