Håvard D. Johansen
Exploring Deep Learning Methods for Real-Time Surgical Instrument Segmentation in Laparoscopy
Aug 03, 2021
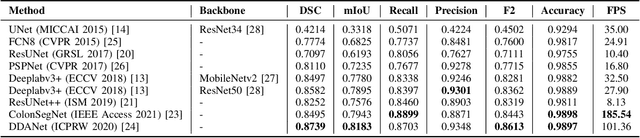
Abstract:Minimally invasive surgery is a surgical intervention used to examine the organs inside the abdomen and has been widely used due to its effectiveness over open surgery. Due to the hardware improvements such as high definition cameras, this procedure has significantly improved and new software methods have demonstrated potential for computer-assisted procedures. However, there exists challenges and requirements to improve detection and tracking of the position of the instruments during these surgical procedures. To this end, we evaluate and compare some popular deep learning methods that can be explored for the automated segmentation of surgical instruments in laparoscopy, an important step towards tool tracking. Our experimental results exhibit that the Dual decoder attention network (DDANet) produces a superior result compared to other recent deep learning methods. DDANet yields a Dice coefficient of 0.8739 and mean intersection-over-union of 0.8183 for the Robust Medical Instrument Segmentation (ROBUST-MIS) Challenge 2019 dataset, at a real-time speed of 101.36 frames-per-second that is critical for such procedures.
A Comprehensive Study on Colorectal Polyp Segmentation with ResUNet++, Conditional Random Field and Test-Time Augmentation
Jul 26, 2021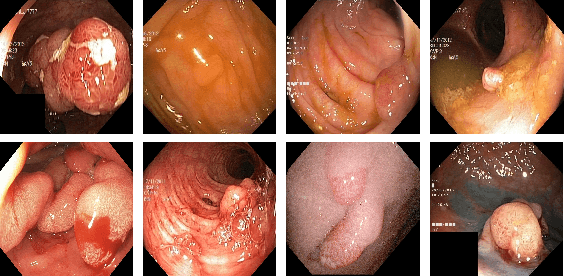
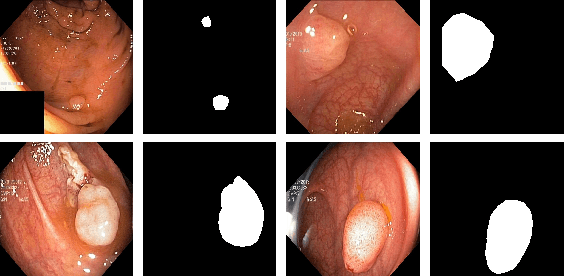
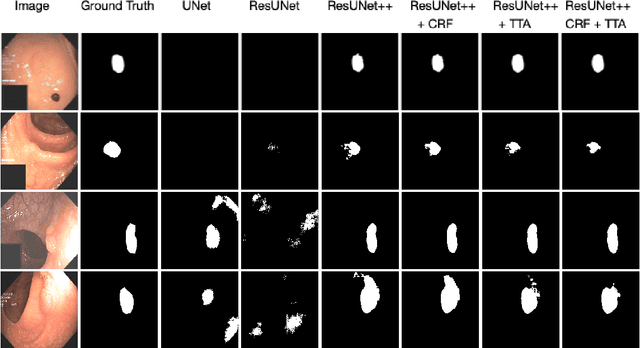
Abstract:Colonoscopy is considered the gold standard for detection of colorectal cancer and its precursors. Existing examination methods are, however, hampered by high overall miss-rate, and many abnormalities are left undetected. Computer-Aided Diagnosis systems based on advanced machine learning algorithms are touted as a game-changer that can identify regions in the colon overlooked by the physicians during endoscopic examinations, and help detect and characterize lesions. In previous work, we have proposed the ResUNet++ architecture and demonstrated that it produces more efficient results compared with its counterparts U-Net and ResUNet. In this paper, we demonstrate that further improvements to the overall prediction performance of the ResUNet++ architecture can be achieved by using conditional random field and test-time augmentation. We have performed extensive evaluations and validated the improvements using six publicly available datasets: Kvasir-SEG, CVC-ClinicDB, CVC-ColonDB, ETIS-Larib Polyp DB, ASU-Mayo Clinic Colonoscopy Video Database, and CVC-VideoClinicDB. Moreover, we compare our proposed architecture and resulting model with other State-of-the-art methods. To explore the generalization capability of ResUNet++ on different publicly available polyp datasets, so that it could be used in a real-world setting, we performed an extensive cross-dataset evaluation. The experimental results show that applying CRF and TTA improves the performance on various polyp segmentation datasets both on the same dataset and cross-dataset.
MSRF-Net: A Multi-Scale Residual Fusion Network for Biomedical Image Segmentation
May 16, 2021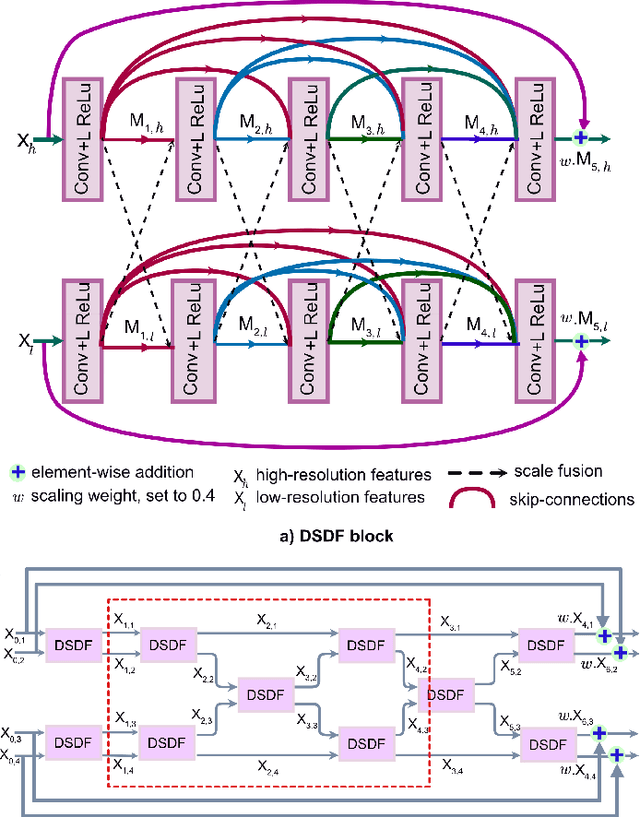
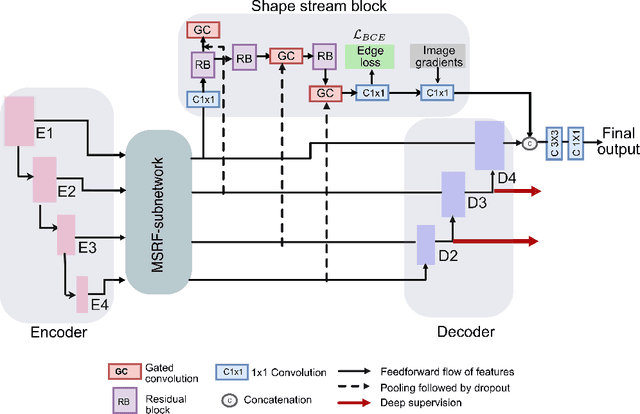
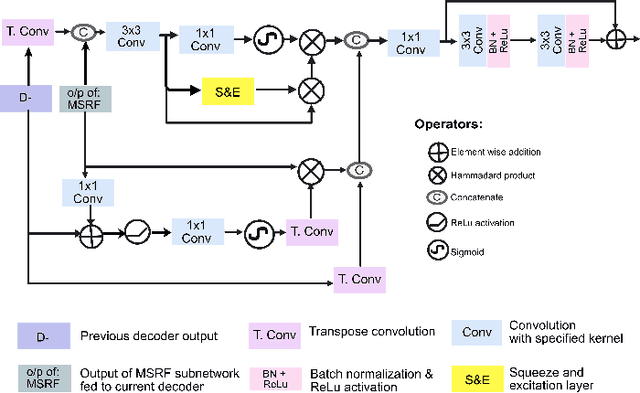
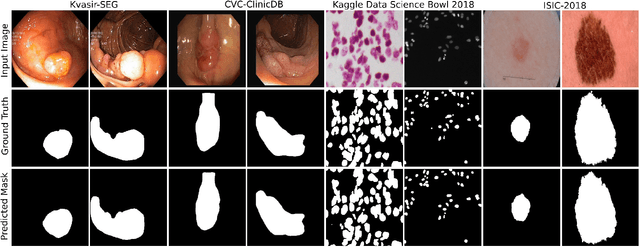
Abstract:Methods based on convolutional neural networks have improved the performance of biomedical image segmentation. However, most of these methods cannot efficiently segment objects of variable sizes and train on small and biased datasets, which are common in biomedical use cases. While methods exist that incorporate multi-scale fusion approaches to address the challenges arising with variable sizes, they usually use complex models that are more suitable for general semantic segmentation computer vision problems. In this paper, we propose a novel architecture called MSRF-Net, which is specially designed for medical image segmentation tasks. The proposed MSRF-Net is able to exchange multi-scale features of varying receptive fields using a dual-scale dense fusion block (DSDF). Our DSDF block can exchange information rigorously across two different resolution scales, and our MSRF sub-network uses multiple DSDF blocks in sequence to perform multi-scale fusion. This allows the preservation of resolution, improved information flow, and propagation of both high- and low-level features to obtain accurate segmentation maps. The proposed MSRF-Net allows to capture object variabilities and provides improved results on different biomedical datasets. Extensive experiments on MSRF-Net demonstrate that the proposed method outperforms most of the cutting-edge medical image segmentation state-of-the-art methods. MSRF-Net advances the performance on four publicly available datasets, and also, MSRF-Net is more generalizable as compared to state-of-the-art methods.
NanoNet: Real-Time Polyp Segmentation in Video Capsule Endoscopy and Colonoscopy
Apr 22, 2021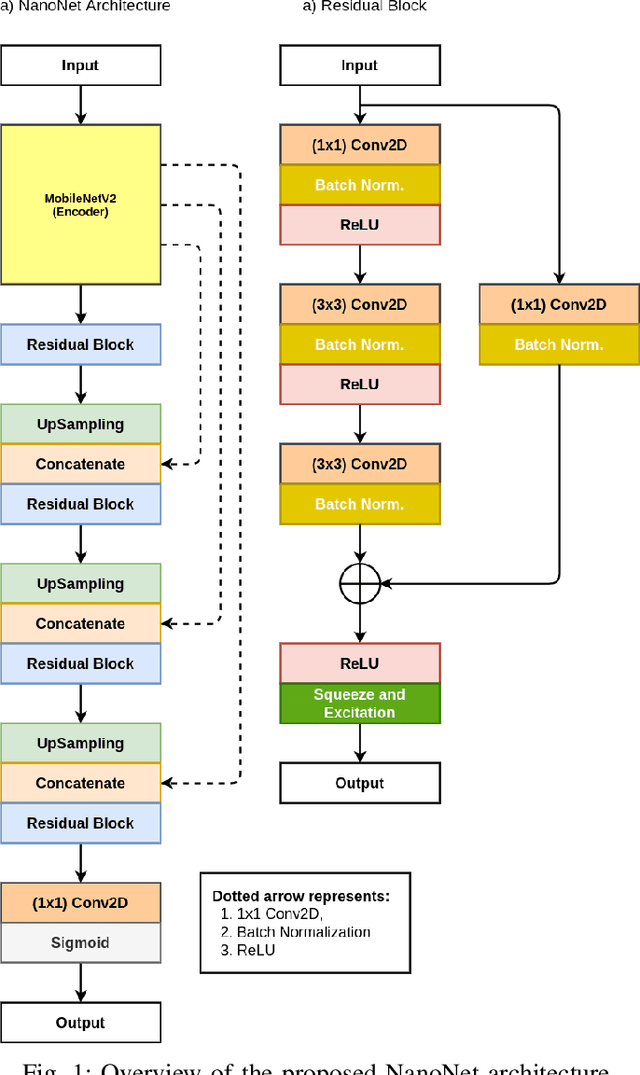
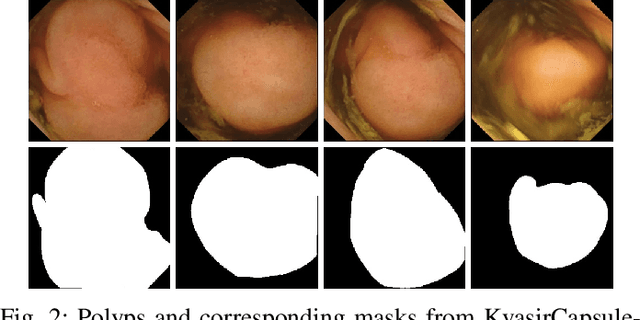
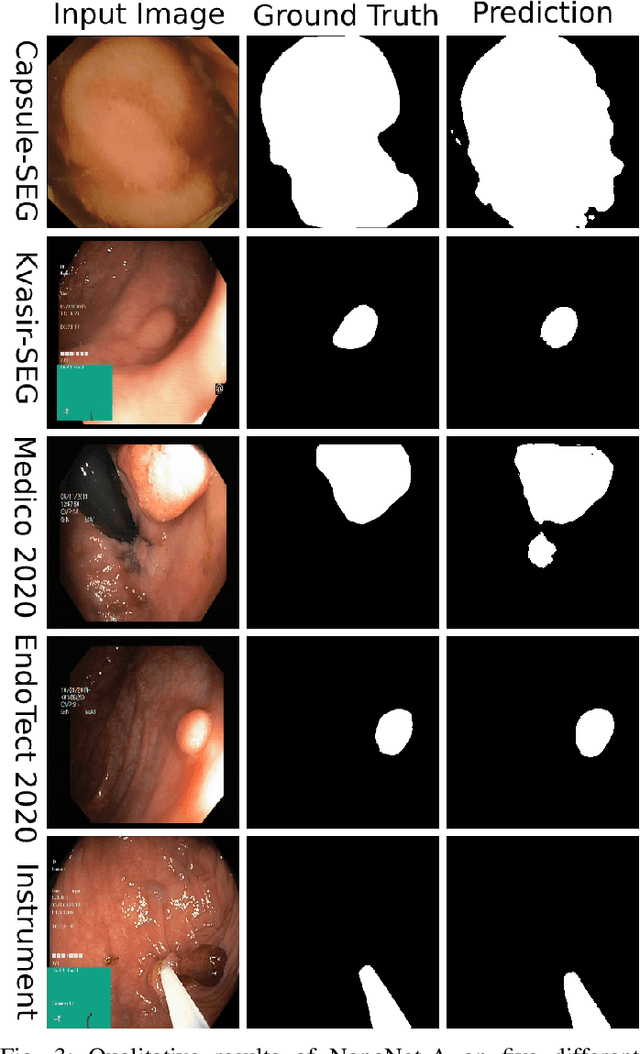
Abstract:Deep learning in gastrointestinal endoscopy can assist to improve clinical performance and be helpful to assess lesions more accurately. To this extent, semantic segmentation methods that can perform automated real-time delineation of a region-of-interest, e.g., boundary identification of cancer or precancerous lesions, can benefit both diagnosis and interventions. However, accurate and real-time segmentation of endoscopic images is extremely challenging due to its high operator dependence and high-definition image quality. To utilize automated methods in clinical settings, it is crucial to design lightweight models with low latency such that they can be integrated with low-end endoscope hardware devices. In this work, we propose NanoNet, a novel architecture for the segmentation of video capsule endoscopy and colonoscopy images. Our proposed architecture allows real-time performance and has higher segmentation accuracy compared to other more complex ones. We use video capsule endoscopy and standard colonoscopy datasets with polyps, and a dataset consisting of endoscopy biopsies and surgical instruments, to evaluate the effectiveness of our approach. Our experiments demonstrate the increased performance of our architecture in terms of a trade-off between model complexity, speed, model parameters, and metric performances. Moreover, the resulting model size is relatively tiny, with only nearly 36,000 parameters compared to traditional deep learning approaches having millions of parameters.
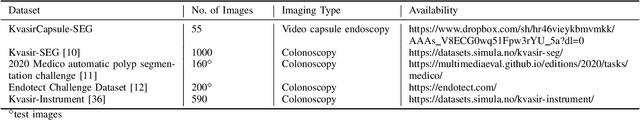
FANet: A Feedback Attention Network for Improved Biomedical Image Segmentation
Mar 31, 2021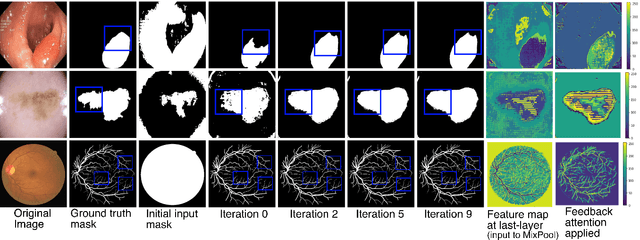
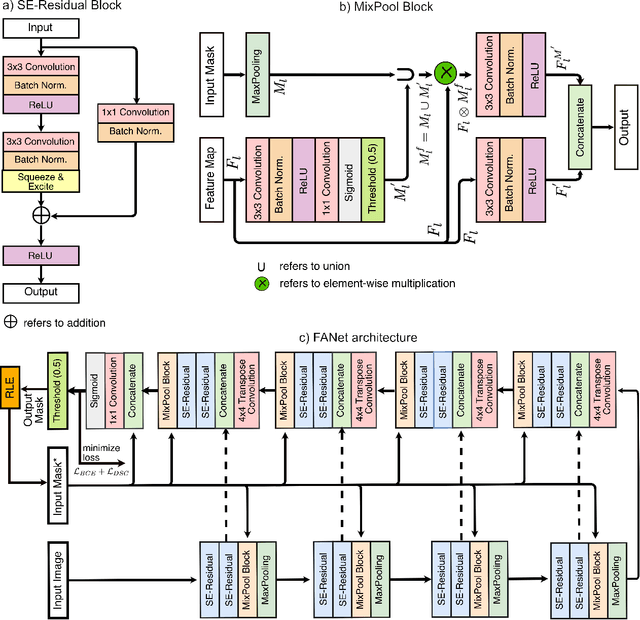
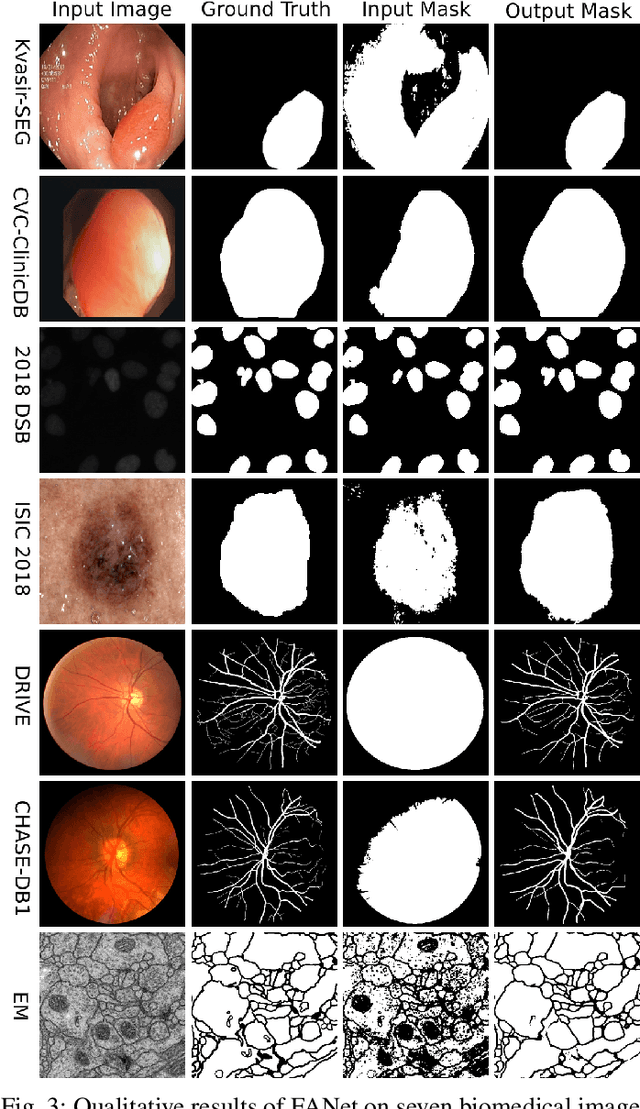
Abstract:With the increase in available large clinical and experimental datasets, there has been substantial amount of work being done on addressing the challenges in the area of biomedical image analysis. Image segmentation, which is crucial for any quantitative analysis, has especially attracted attention. Recent hardware advancement has led to the success of deep learning approaches. However, although deep learning models are being trained on large datasets, existing methods do not use the information from different learning epochs effectively. In this work, we leverage the information of each training epoch to prune the prediction maps of the subsequent epochs. We propose a novel architecture called feedback attention network (FANet) that unifies the previous epoch mask with the feature map of the current training epoch. The previous epoch mask is then used to provide a hard attention to the learnt feature maps at different convolutional layers. The network also allows to rectify the predictions in an iterative fashion during the test time. We show that our proposed feedback attention model provides a substantial improvement on most segmentation metrics tested on seven publicly available biomedical imaging datasets demonstrating the effectiveness of the proposed FANet.
LightLayers: Parameter Efficient Dense and Convolutional Layers for Image Classification
Jan 06, 2021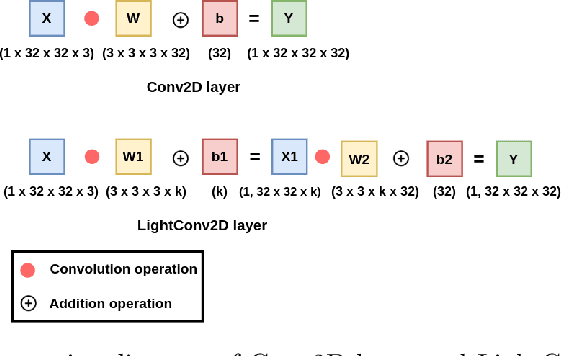
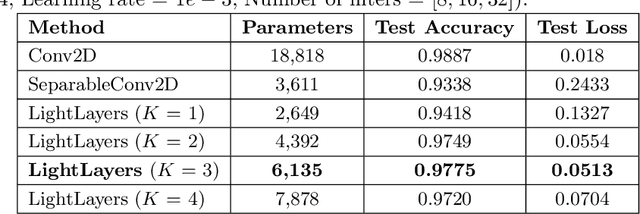
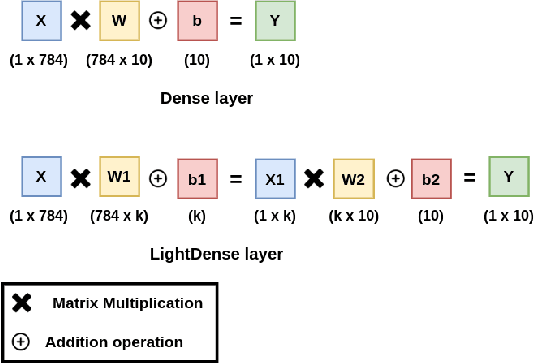

Abstract:Deep Neural Networks (DNNs) have become the de-facto standard in computer vision, as well as in many other pattern recognition tasks. A key drawback of DNNs is that the training phase can be very computationally expensive. Organizations or individuals that cannot afford purchasing state-of-the-art hardware or tapping into cloud-hosted infrastructures may face a long waiting time before the training completes or might not be able to train a model at all. Investigating novel ways to reduce the training time could be a potential solution to alleviate this drawback, and thus enabling more rapid development of new algorithms and models. In this paper, we propose LightLayers, a method for reducing the number of trainable parameters in deep neural networks (DNN). The proposed LightLayers consists of LightDense andLightConv2D layer that are as efficient as regular Conv2D and Dense layers, but uses less parameters. We resort to Matrix Factorization to reduce the complexity of the DNN models resulting into lightweight DNNmodels that require less computational power, without much loss in the accuracy. We have tested LightLayers on MNIST, Fashion MNIST, CI-FAR 10, and CIFAR 100 datasets. Promising results are obtained for MNIST, Fashion MNIST, CIFAR-10 datasets whereas CIFAR 100 shows acceptable performance by using fewer parameters.
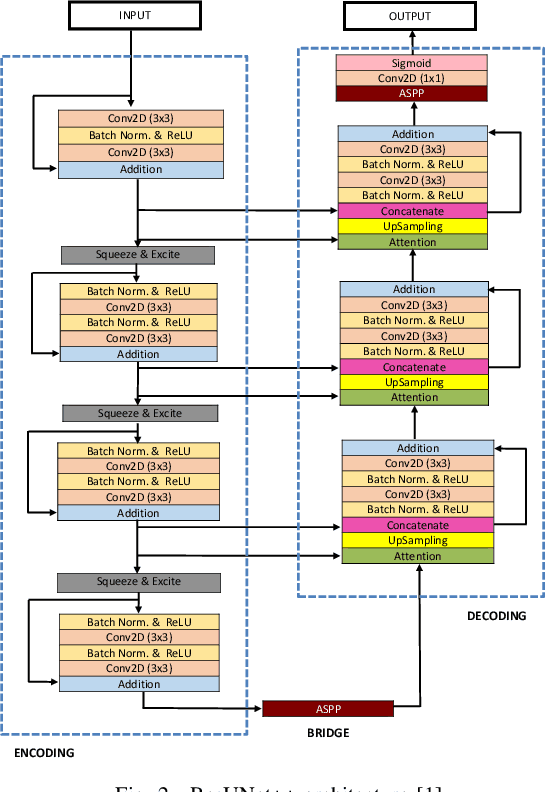
DDANet: Dual Decoder Attention Network for Automatic Polyp Segmentation
Dec 30, 2020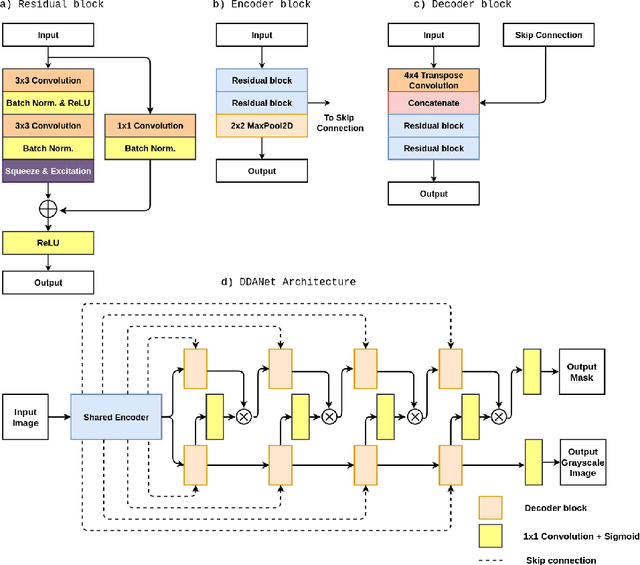
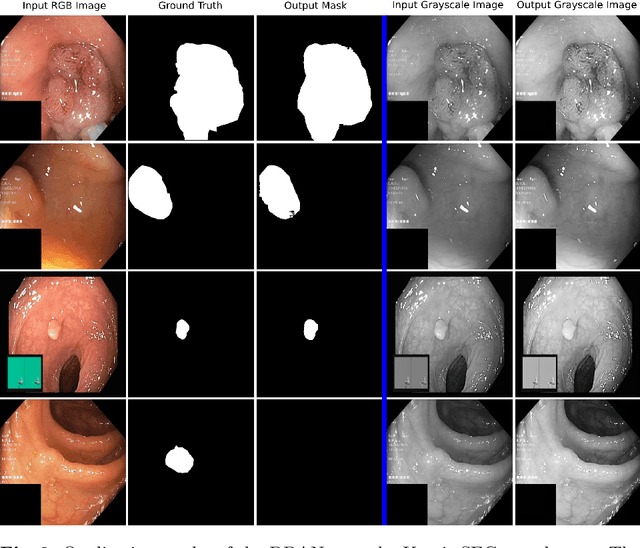
Abstract:Colonoscopy is the gold standard for examination and detection of colorectal polyps. Localization and delineation of polyps can play a vital role in treatment (e.g., surgical planning) and prognostic decision making. Polyp segmentation can provide detailed boundary information for clinical analysis. Convolutional neural networks have improved the performance in colonoscopy. However, polyps usually possess various challenges, such as intra-and inter-class variation and noise. While manual labeling for polyp assessment requires time from experts and is prone to human error (e.g., missed lesions), an automated, accurate, and fast segmentation can improve the quality of delineated lesion boundaries and reduce missed rate. The Endotect challenge provides an opportunity to benchmark computer vision methods by training on the publicly available Hyperkvasir and testing on a separate unseen dataset. In this paper, we propose a novel architecture called ``DDANet'' based on a dual decoder attention network. Our experiments demonstrate that the model trained on the Kvasir-SEG dataset and tested on an unseen dataset achieves a dice coefficient of 0.7874, mIoU of 0.7010, recall of 0.7987, and a precision of 0.8577, demonstrating the generalization ability of our model.
Real-Time Polyp Detection, Localisation and Segmentation in Colonoscopy Using Deep Learning
Nov 15, 2020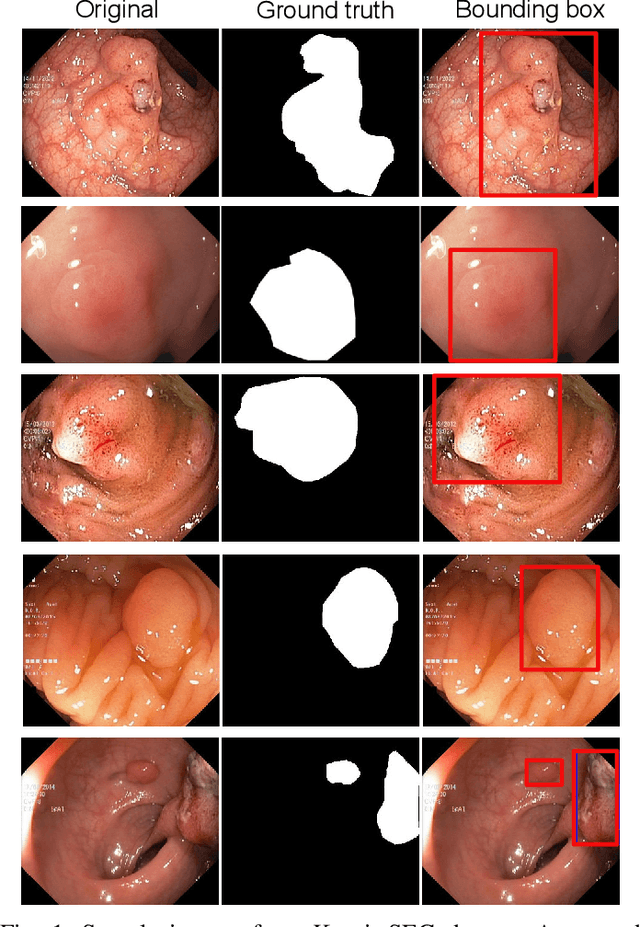
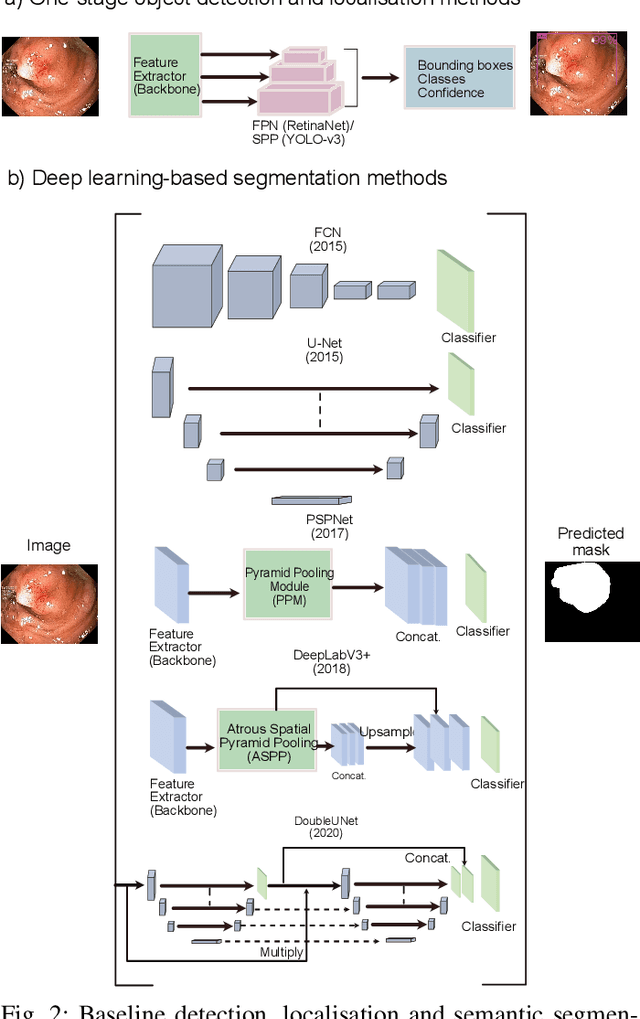


Abstract:Computer-aided detection, localisation, and segmentation methods can help improve colonoscopy procedures. Even though many methods have been built to tackle automatic detection and segmentation of polyps, benchmarking of state-of-the-art methods still remains an open problem. This is due to the increasing number of researched computer-vision methods that can be applied to polyp datasets. Benchmarking of novel methods can provide a direction to the development of automated polyp detection and segmentation tasks. Furthermore, it ensures that the produced results in the community are reproducible and provide a fair comparison of developed methods. In this paper, we benchmark several recent state-of-the-art methods using Kvasir-SEG, an open-access dataset of colonoscopy images, for polyp detection, localisation, and segmentation evaluating both method accuracy and speed. Whilst, most methods in literature have competitive performance over accuracy, we show that YOLOv4 with a Darknet53 backbone and cross-stage-partial connections achieved a better trade-off between an average precision of 0.8513 and mean IoU of 0.8025, and the fastest speed of 48 frames per second for the detection and localisation task. Likewise, UNet with a ResNet34 backbone achieved the highest dice coefficient of 0.8757 and the best average speed of 35 frames per second for the segmentation task. Our comprehensive comparison with various state-of-the-art methods reveal the importance of benchmarking the deep learning methods for automated real-time polyp identification and delineations that can potentially transform current clinical practices and minimise miss-detection rates.
DoubleU-Net: A Deep Convolutional Neural Network for Medical Image Segmentation
Jun 27, 2020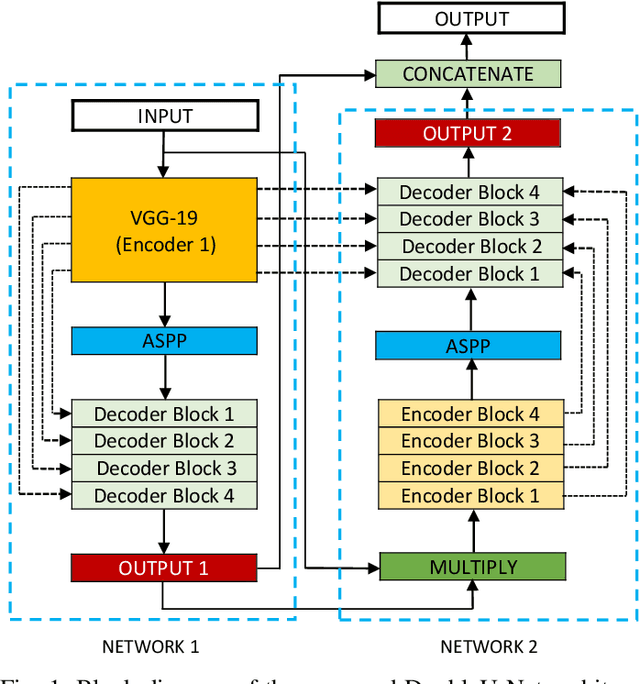



Abstract:Semantic image segmentation is the process of labeling each pixel of an image with its corresponding class. An encoder-decoder based approach, like U-Net and its variants, is a popular strategy for solving medical image segmentation tasks. To improve the performance of U-Net on various segmentation tasks, we propose a novel architecture called DoubleU-Net, which is a combination of two U-Net architectures stacked on top of each other. The first U-Net uses a pre-trained VGG-19 as the encoder, which has already learned features from ImageNet and can be transferred to another task easily. To capture more semantic information efficiently, we added another U-Net at the bottom. We also adopt Atrous Spatial Pyramid Pooling (ASPP) to capture contextual information within the network. We have evaluated DoubleU-Net using four medical segmentation datasets, covering various imaging modalities such as colonoscopy, dermoscopy, and microscopy. Experiments on the MICCAI 2015 segmentation challenge, the CVC-ClinicDB, the 2018 Data Science Bowl challenge, and the Lesion boundary segmentation datasets demonstrate that the DoubleU-Net outperforms U-Net and the baseline models. Moreover, DoubleU-Net produces more accurate segmentation masks, especially in the case of the CVC-ClinicDB and MICCAI 2015 segmentation challenge datasets, which have challenging images such as smaller and flat polyps. These results show the improvement over the existing U-Net model. The encouraging results, produced on various medical image segmentation datasets, show that DoubleU-Net can be used as a strong baseline for both medical image segmentation and cross-dataset evaluation testing to measure the generalizability of Deep Learning (DL) models.
An Extensive Study on Cross-Dataset Bias and Evaluation Metrics Interpretation for Machine Learning applied to Gastrointestinal Tract Abnormality Classification
May 08, 2020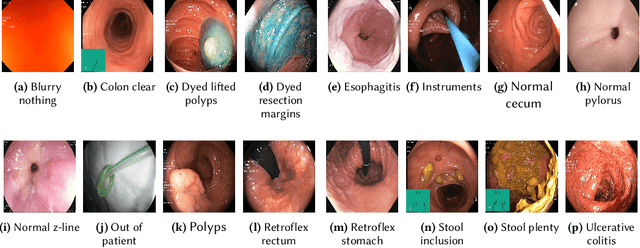
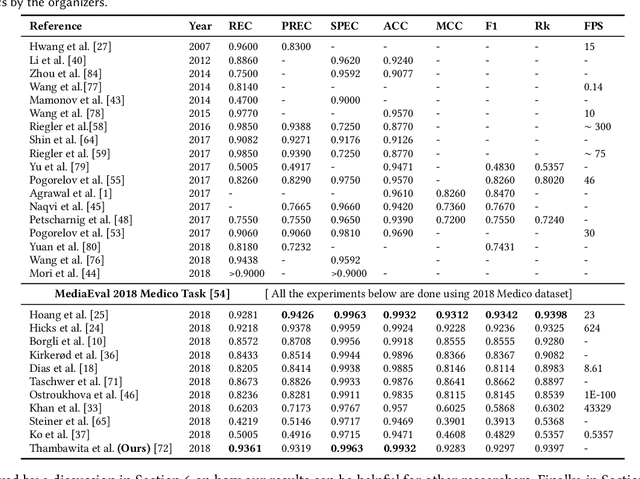
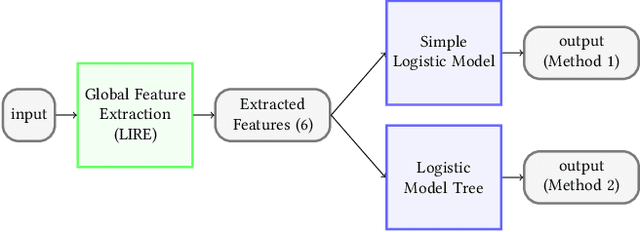

Abstract:Precise and efficient automated identification of Gastrointestinal (GI) tract diseases can help doctors treat more patients and improve the rate of disease detection and identification. Currently, automatic analysis of diseases in the GI tract is a hot topic in both computer science and medical-related journals. Nevertheless, the evaluation of such an automatic analysis is often incomplete or simply wrong. Algorithms are often only tested on small and biased datasets, and cross-dataset evaluations are rarely performed. A clear understanding of evaluation metrics and machine learning models with cross datasets is crucial to bring research in the field to a new quality level. Towards this goal, we present comprehensive evaluations of five distinct machine learning models using Global Features and Deep Neural Networks that can classify 16 different key types of GI tract conditions, including pathological findings, anatomical landmarks, polyp removal conditions, and normal findings from images captured by common GI tract examination instruments. In our evaluation, we introduce performance hexagons using six performance metrics such as recall, precision, specificity, accuracy, F1-score, and Matthews Correlation Coefficient to demonstrate how to determine the real capabilities of models rather than evaluating them shallowly. Furthermore, we perform cross-dataset evaluations using different datasets for training and testing. With these cross-dataset evaluations, we demonstrate the challenge of actually building a generalizable model that could be used across different hospitals. Our experiments clearly show that more sophisticated performance metrics and evaluation methods need to be applied to get reliable models rather than depending on evaluations of the splits of the same dataset, i.e., the performance metrics should always be interpreted together rather than relying on a single metric.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge