Gamze Gursoy
KEEP: Integrating Medical Ontologies with Clinical Data for Robust Code Embeddings
Oct 06, 2025Abstract:Machine learning in healthcare requires effective representation of structured medical codes, but current methods face a trade off: knowledge graph based approaches capture formal relationships but miss real world patterns, while data driven methods learn empirical associations but often overlook structured knowledge in medical terminologies. We present KEEP (Knowledge preserving and Empirically refined Embedding Process), an efficient framework that bridges this gap by combining knowledge graph embeddings with adaptive learning from clinical data. KEEP first generates embeddings from knowledge graphs, then employs regularized training on patient records to adaptively integrate empirical patterns while preserving ontological relationships. Importantly, KEEP produces final embeddings without task specific auxiliary or end to end training enabling KEEP to support multiple downstream applications and model architectures. Evaluations on structured EHR from UK Biobank and MIMIC IV demonstrate that KEEP outperforms both traditional and Language Model based approaches in capturing semantic relationships and predicting clinical outcomes. Moreover, KEEP's minimal computational requirements make it particularly suitable for resource constrained environments.
A Universal Metric of Dataset Similarity for Cross-silo Federated Learning
Apr 29, 2024Abstract:Federated Learning is increasingly used in domains such as healthcare to facilitate collaborative model training without data-sharing. However, datasets located in different sites are often non-identically distributed, leading to degradation of model performance in FL. Most existing methods for assessing these distribution shifts are limited by being dataset or task-specific. Moreover, these metrics can only be calculated by exchanging data, a practice restricted in many FL scenarios. To address these challenges, we propose a novel metric for assessing dataset similarity. Our metric exhibits several desirable properties for FL: it is dataset-agnostic, is calculated in a privacy-preserving manner, and is computationally efficient, requiring no model training. In this paper, we first establish a theoretical connection between our metric and training dynamics in FL. Next, we extensively evaluate our metric on a range of datasets including synthetic, benchmark, and medical imaging datasets. We demonstrate that our metric shows a robust and interpretable relationship with model performance and can be calculated in privacy-preserving manner. As the first federated dataset similarity metric, we believe this metric can better facilitate successful collaborations between sites.
Recent Advances, Applications, and Open Challenges in Machine Learning for Health: Reflections from Research Roundtables at ML4H 2023 Symposium
Mar 03, 2024Abstract:The third ML4H symposium was held in person on December 10, 2023, in New Orleans, Louisiana, USA. The symposium included research roundtable sessions to foster discussions between participants and senior researchers on timely and relevant topics for the \ac{ML4H} community. Encouraged by the successful virtual roundtables in the previous year, we organized eleven in-person roundtables and four virtual roundtables at ML4H 2022. The organization of the research roundtables at the conference involved 17 Senior Chairs and 19 Junior Chairs across 11 tables. Each roundtable session included invited senior chairs (with substantial experience in the field), junior chairs (responsible for facilitating the discussion), and attendees from diverse backgrounds with interest in the session's topic. Herein we detail the organization process and compile takeaways from these roundtable discussions, including recent advances, applications, and open challenges for each topic. We conclude with a summary and lessons learned across all roundtables. This document serves as a comprehensive review paper, summarizing the recent advancements in machine learning for healthcare as contributed by foremost researchers in the field.
Privacy-preserving patient clustering for personalized federated learning
Jul 17, 2023Abstract:Federated Learning (FL) is a machine learning framework that enables multiple organizations to train a model without sharing their data with a central server. However, it experiences significant performance degradation if the data is non-identically independently distributed (non-IID). This is a problem in medical settings, where variations in the patient population contribute significantly to distribution differences across hospitals. Personalized FL addresses this issue by accounting for site-specific distribution differences. Clustered FL, a Personalized FL variant, was used to address this problem by clustering patients into groups across hospitals and training separate models on each group. However, privacy concerns remained as a challenge as the clustering process requires exchange of patient-level information. This was previously solved by forming clusters using aggregated data, which led to inaccurate groups and performance degradation. In this study, we propose Privacy-preserving Community-Based Federated machine Learning (PCBFL), a novel Clustered FL framework that can cluster patients using patient-level data while protecting privacy. PCBFL uses Secure Multiparty Computation, a cryptographic technique, to securely calculate patient-level similarity scores across hospitals. We then evaluate PCBFL by training a federated mortality prediction model using 20 sites from the eICU dataset. We compare the performance gain from PCBFL against traditional and existing Clustered FL frameworks. Our results show that PCBFL successfully forms clinically meaningful cohorts of low, medium, and high-risk patients. PCBFL outperforms traditional and existing Clustered FL frameworks with an average AUC improvement of 4.3% and AUPRC improvement of 7.8%.
Scalable privacy-preserving cancer type prediction with homomorphic encryption
Apr 12, 2022
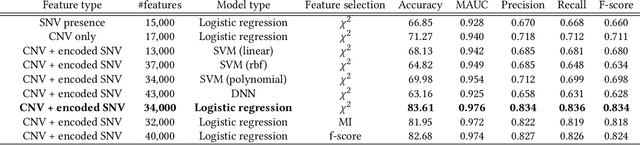
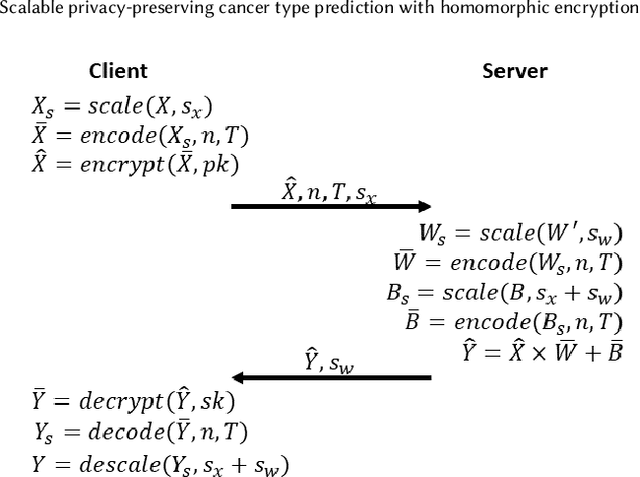
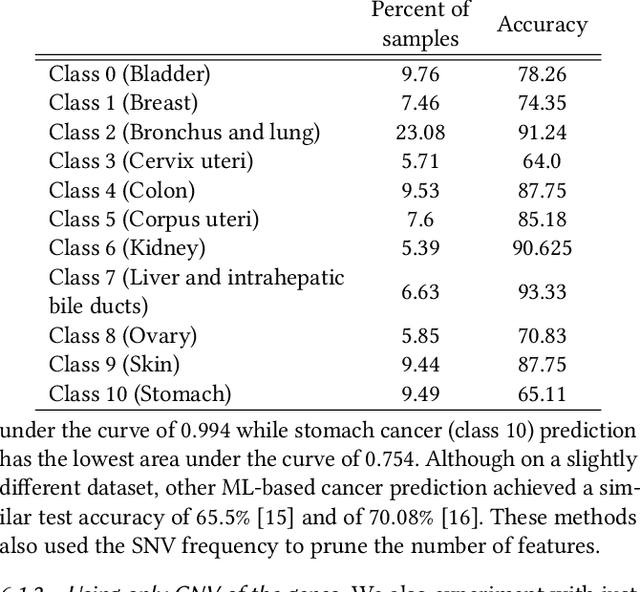
Abstract:Machine Learning (ML) alleviates the challenges of high-dimensional data analysis and improves decision making in critical applications like healthcare. Effective cancer type from high-dimensional genetic mutation data can be useful for cancer diagnosis and treatment, if the distinguishable patterns between cancer types are identified. At the same time, analysis of high-dimensional data is computationally expensive and is often outsourced to cloud services. Privacy concerns in outsourced ML, especially in the field of genetics, motivate the use of encrypted computation, like Homomorphic Encryption (HE). But restrictive overheads of encrypted computation deter its usage. In this work, we explore the challenges of privacy preserving cancer detection using a real-world dataset consisting of more than 2 million genetic information for several cancer types. Since the data is inherently high-dimensional, we explore smaller ML models for cancer prediction to enable fast inference in the privacy preserving domain. We develop a solution for privacy preserving cancer inference which first leverages the domain knowledge on somatic mutations to efficiently encode genetic mutations and then uses statistical tests for feature selection. Our logistic regression model, built using our novel encoding scheme, achieves 0.98 micro-average area under curve with 13% higher test accuracy than similar studies. We exhaustively test our model's predictive capabilities by analyzing the genes used by the model. Furthermore, we propose a fast matrix multiplication algorithm that can efficiently handle high-dimensional data. Experimental results show that, even with 40,000 features, our proposed matrix multiplication algorithm can speed up concurrent inference of multiple individuals by approximately 10x and inference of a single individual by approximately 550x, in comparison to standard matrix multiplication.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge