Eugene W. Myers
Estimation of Optical Aberrations in 3D Microscopic Bioimages
Sep 16, 2022



Abstract:The quality of microscopy images often suffers from optical aberrations. These aberrations and their associated point spread functions have to be quantitatively estimated to restore aberrated images. The recent state-of-the-art method PhaseNet, based on a convolutional neural network, can quantify aberrations accurately but is limited to images of point light sources, e.g. fluorescent beads. In this research, we describe an extension of PhaseNet enabling its use on 3D images of biological samples. To this end, our method incorporates object-specific information into the simulated images used for training the network. Further, we add a Python-based restoration of images via Richardson-Lucy deconvolution. We demonstrate that the deconvolution with the predicted PSF can not only remove the simulated aberrations but also improve the quality of the real raw microscopic images with unknown residual PSF. We provide code for fast and convenient prediction and correction of aberrations.
Fast and accurate aberration estimation from 3D bead images using convolutional neural networks
Jun 02, 2020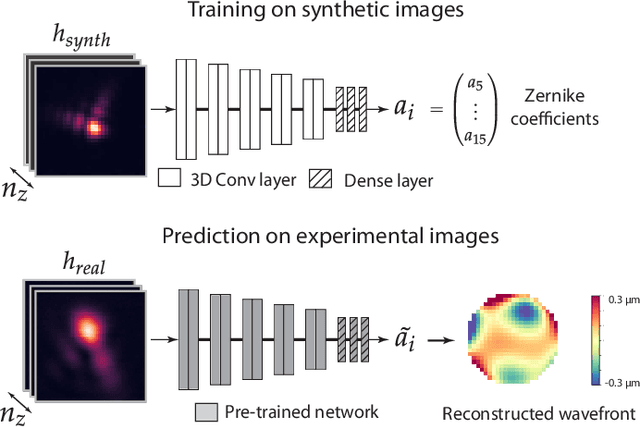

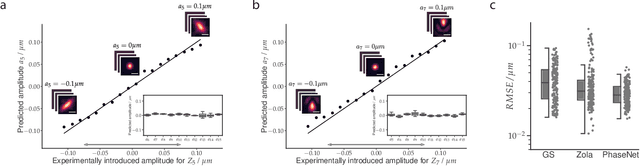
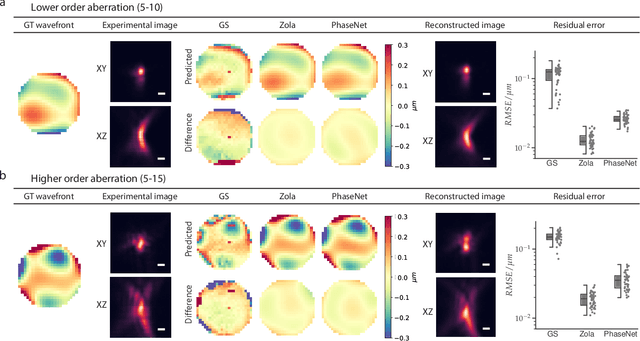
Abstract:Estimating optical aberrations from volumetric intensity images is a key step in sensorless adaptive optics for microscopy. Here we describe a method (PHASENET) for fast and accurate aberration measurement from experimentally acquired 3D bead images using convolutional neural networks. Importantly, we show that networks trained only on synthetically generated data can successfully predict aberrations from experimental images. We demonstrate our approach on two data sets acquired with different microscopy modalities and find that PHASENET yields results better than or comparable to classical methods while being orders of magnitude faster. We furthermore show that the number of focal planes required for satisfactory prediction is related to different symmetry groups of Zernike modes. PHASENET is freely available as open-source software in Python.
Mapping Auto-context Decision Forests to Deep ConvNets for Semantic Segmentation
Aug 13, 2018



Abstract:We consider the task of pixel-wise semantic segmentation given a small set of labeled training images. Among two of the most popular techniques to address this task are Decision Forests (DF) and Neural Networks (NN). In this work, we explore the relationship between two special forms of these techniques: stacked DFs (namely Auto-context) and deep Convolutional Neural Networks (ConvNet). Our main contribution is to show that Auto-context can be mapped to a deep ConvNet with novel architecture, and thereby trained end-to-end. This mapping can be used as an initialization of a deep ConvNet, enabling training even in the face of very limited amounts of training data. We also demonstrate an approximate mapping back from the refined ConvNet to a second stacked DF, with improved performance over the original. We experimentally verify that these mappings outperform stacked DFs for two different applications in computer vision and biology: Kinect-based body part labeling from depth images, and somite segmentation in microscopy images of developing zebrafish. Finally, we revisit the core mapping from a Decision Tree (DT) to a NN, and show that it is also possible to map a fuzzy DT, with sigmoidal split decisions, to a NN. This addresses multiple limitations of the previous mapping, and yields new insights into the popular Rectified Linear Unit (ReLU), and more recently proposed concatenated ReLU (CReLU), activation functions.
Efficient Algorithms for Moral Lineage Tracing
Aug 25, 2017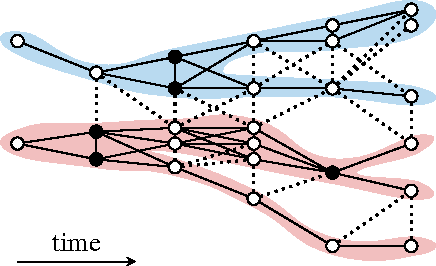
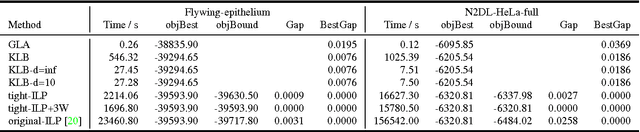
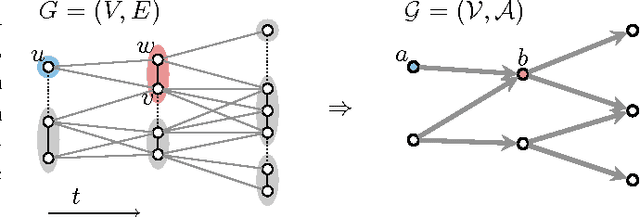
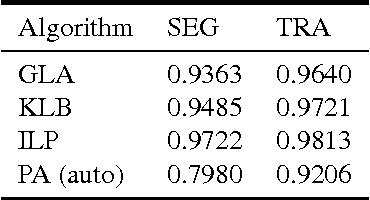
Abstract:Lineage tracing, the joint segmentation and tracking of living cells as they move and divide in a sequence of light microscopy images, is a challenging task. Jug et al. have proposed a mathematical abstraction of this task, the moral lineage tracing problem (MLTP), whose feasible solutions define both a segmentation of every image and a lineage forest of cells. Their branch-and-cut algorithm, however, is prone to many cuts and slow convergence for large instances. To address this problem, we make three contributions: (i) we devise the first efficient primal feasible local search algorithms for the MLTP, (ii) we improve the branch-and-cut algorithm by separating tighter cutting planes and by incorporating our primal algorithms, (iii) we show in experiments that our algorithms find accurate solutions on the problem instances of Jug et al. and scale to larger instances, leveraging moral lineage tracing to practical significance.
Moral Lineage Tracing
Nov 08, 2016



Abstract:Lineage tracing, the tracking of living cells as they move and divide, is a central problem in biological image analysis. Solutions, called lineage forests, are key to understanding how the structure of multicellular organisms emerges. We propose an integer linear program (ILP) whose feasible solutions define a decomposition of each image in a sequence into cells (segmentation), and a lineage forest of cells across images (tracing). Unlike previous formulations, we do not constrain the set of decompositions, except by contracting pixels to superpixels. The main challenge, as we show, is to enforce the morality of lineages, i.e., the constraint that cells do not merge. To enforce morality, we introduce path-cut inequalities. To find feasible solutions of the NP-hard ILP, with certified bounds to the global optimum, we define efficient separation procedures and apply these as part of a branch-and-cut algorithm. We show the effectiveness of this approach by analyzing feasible solutions for real microscopy data in terms of bounds and run-time, and by their weighted edit distance to ground truth lineage forests traced by humans.
* 11 pages
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge