Emanuele Olivetti
Supervised Tractogram Filtering using Geometric Deep Learning
Dec 06, 2022



Abstract:A tractogram is a virtual representation of the brain white matter. It is composed of millions of virtual fibers, encoded as 3D polylines, which approximate the white matter axonal pathways. To date, tractograms are the most accurate white matter representation and thus are used for tasks like presurgical planning and investigations of neuroplasticity, brain disorders, or brain networks. However, it is a well-known issue that a large portion of tractogram fibers is not anatomically plausible and can be considered artifacts of the tracking procedure. With Verifyber, we tackle the problem of filtering out such non-plausible fibers using a novel fully-supervised learning approach. Differently from other approaches based on signal reconstruction and/or brain topology regularization, we guide our method with the existing anatomical knowledge of the white matter. Using tractograms annotated according to anatomical principles, we train our model, Verifyber, to classify fibers as either anatomically plausible or non-plausible. The proposed Verifyber model is an original Geometric Deep Learning method that can deal with variable size fibers, while being invariant to fiber orientation. Our model considers each fiber as a graph of points, and by learning features of the edges between consecutive points via the proposed sequence Edge Convolution, it can capture the underlying anatomical properties. The output filtering results highly accurate and robust across an extensive set of experiments, and fast; with a 12GB GPU, filtering a tractogram of 1M fibers requires less than a minute. Verifyber implementation and trained models are available at https://github.com/FBK-NILab/verifyber.
Tractogram filtering of anatomically non-plausible fibers with geometric deep learning
Mar 24, 2020



Abstract:Tractograms are virtual representations of the white matter fibers of the brain. They are of primary interest for tasks like presurgical planning, and investigation of neuroplasticity or brain disorders. Each tractogram is composed of millions of fibers encoded as 3D polylines. Unfortunately, a large portion of those fibers are not anatomically plausible and can be considered artifacts of the tracking algorithms. Common methods for tractogram filtering are based on signal reconstruction, a principled approach, but unable to consider the knowledge of brain anatomy. In this work, we address the problem of tractogram filtering as a supervised learning problem by exploiting the ground truth annotations obtained with a recent heuristic method, which labels fibers as either anatomically plausible or non-plausible according to well-established anatomical properties. The intuitive idea is to model a fiber as a point cloud and the goal is to investigate whether and how a geometric deep learning model might capture its anatomical properties. Our contribution is an extension of the Dynamic Edge Convolution model that exploits the sequential relations of points in a fiber and discriminates with high accuracy plausible/non-plausible fibers.
A Test for Shared Patterns in Cross-modal Brain Activation Analysis
Oct 08, 2019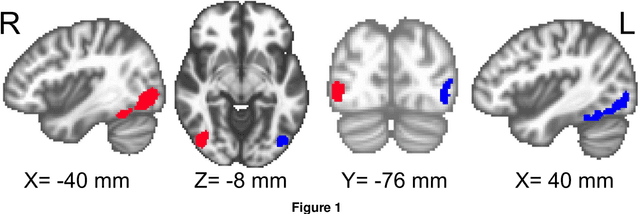
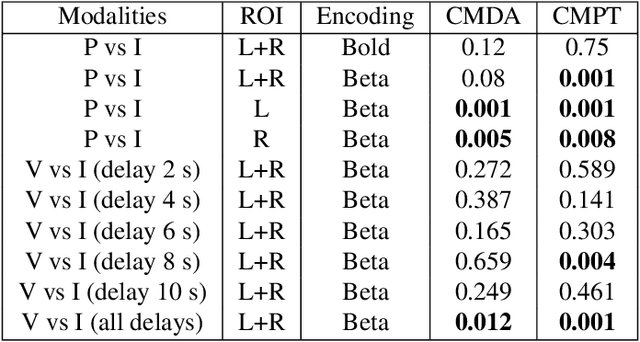
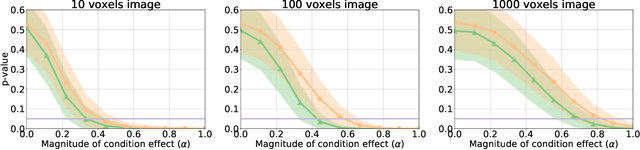
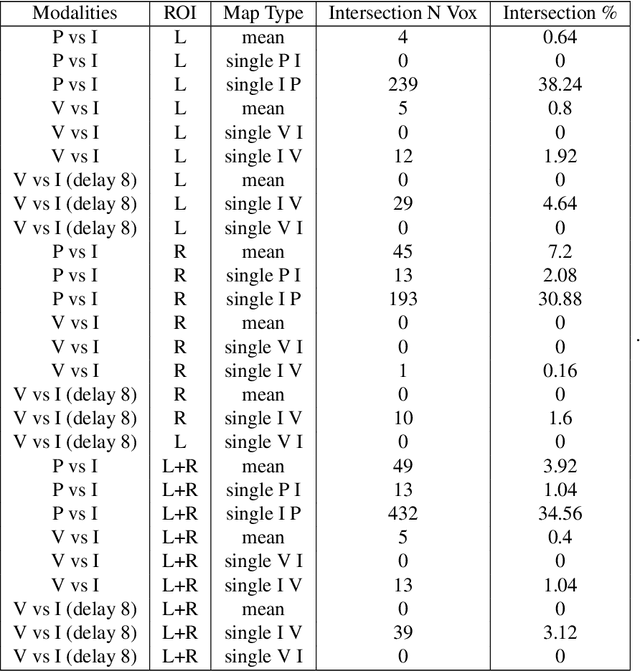
Abstract:Determining the extent to which different cognitive modalities (understood here as the set of cognitive processes underlying the elaboration of a stimulus by the brain) rely on overlapping neural representations is a fundamental issue in cognitive neuroscience. In the last decade, the identification of shared activity patterns has been mostly framed as a supervised learning problem. For instance, a classifier is trained to discriminate categories (e.g. faces vs. houses) in modality I (e.g. perception) and tested on the same categories in modality II (e.g. imagery). This type of analysis is often referred to as cross-modal decoding. In this paper we take a different approach and instead formulate the problem of assessing shared patterns across modalities within the framework of statistical hypothesis testing. We propose both an appropriate test statistic and a scheme based on permutation testing to compute the significance of this test while making only minimal distributional assumption. We denote this test cross-modal permutation test (CMPT). We also provide empirical evidence on synthetic datasets that our approach has greater statistical power than the cross-modal decoding method while maintaining low Type I errors (rejecting a true null hypothesis). We compare both approaches on an fMRI dataset with three different cognitive modalities (perception, imagery, visual search). Finally, we show how CMPT can be combined with Searchlight analysis to explore spatial distribution of shared activity patterns.
Anatomically-Informed Multiple Linear Assignment Problems for White Matter Bundle Segmentation
Jul 16, 2019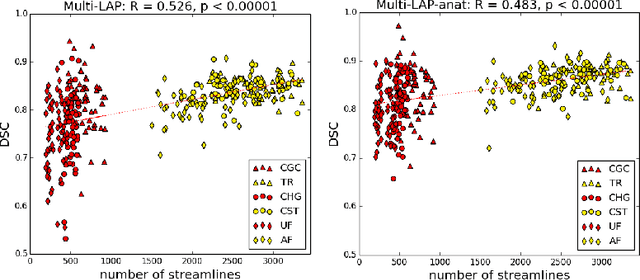

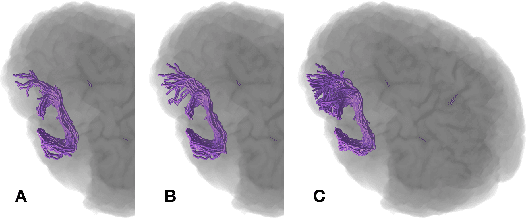
Abstract:Segmenting white matter bundles from human tractograms is a task of interest for several applications. Current methods for bundle segmentation consider either only prior knowledge about the relative anatomical position of a bundle, or only its geometrical properties. Our aim is to improve the results of segmentation by proposing a method that takes into account information about both the underlying anatomy and the geometry of bundles at the same time. To achieve this goal, we extend a state-of-the-art example-based method based on the Linear Assignment Problem (LAP) by including prior anatomical information within the optimization process. The proposed method shows a significant improvement with respect to the original method, in particular on small bundles.
Comparison of Distances for Supervised Segmentation of White Matter Tractography
Aug 04, 2017


Abstract:Tractograms are mathematical representations of the main paths of axons within the white matter of the brain, from diffusion MRI data. Such representations are in the form of polylines, called streamlines, and one streamline approximates the common path of tens of thousands of axons. The analysis of tractograms is a task of interest in multiple fields, like neurosurgery and neurology. A basic building block of many pipelines of analysis is the definition of a distance function between streamlines. Multiple distance functions have been proposed in the literature, and different authors use different distances, usually without a specific reason other than invoking the "common practice". To this end, in this work we want to test such common practices, in order to obtain factual reasons for choosing one distance over another. For these reasons, in this work we compare many streamline distance functions available in the literature. We focus on the common task of automatic bundle segmentation and we adopt the recent approach of supervised segmentation from expert-based examples. Using the HCP dataset, we compare several distances obtaining guidelines on the choice of which distance function one should use for supervised bundle segmentation.
Mapping Tractography Across Subjects
Jan 29, 2016


Abstract:Diffusion magnetic resonance imaging (dMRI) and tractography provide means to study the anatomical structures within the white matter of the brain. When studying tractography data across subjects, it is usually necessary to align, i.e. to register, tractographies together. This registration step is most often performed by applying the transformation resulting from the registration of other volumetric images (T1, FA). In contrast with registration methods that "transform" tractographies, in this work, we try to find which streamline in one tractography correspond to which streamline in the other tractography, without any transformation. In other words, we try to find a "mapping" between the tractographies. We propose a graph-based solution for the tractography mapping problem and we explain similarities and differences with the related well-known graph matching problem. Specifically, we define a loss function based on the pairwise streamline distance and reformulate the mapping problem as combinatorial optimization of that loss function. We show preliminary promising results where we compare the proposed method, implemented with simulated annealing, against a standard registration techniques in a task of segmentation of the corticospinal tract.
The Kernel Two-Sample Test for Brain Networks
Nov 19, 2015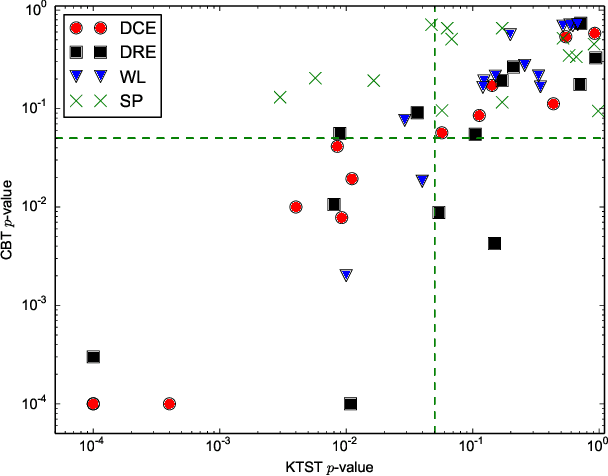



Abstract:In clinical and neuroscientific studies, systematic differences between two populations of brain networks are investigated in order to characterize mental diseases or processes. Those networks are usually represented as graphs built from neuroimaging data and studied by means of graph analysis methods. The typical machine learning approach to study these brain graphs creates a classifier and tests its ability to discriminate the two populations. In contrast to this approach, in this work we propose to directly test whether two populations of graphs are different or not, by using the kernel two-sample test (KTST), without creating the intermediate classifier. We claim that, in general, the two approaches provides similar results and that the KTST requires much less computation. Additionally, in the regime of low sample size, we claim that the KTST has lower frequency of Type II error than the classification approach. Besides providing algorithmic considerations to support these claims, we show strong evidence through experiments and one simulation.
The Approximation of the Dissimilarity Projection
Apr 02, 2015



Abstract:Diffusion magnetic resonance imaging (dMRI) data allow to reconstruct the 3D pathways of axons within the white matter of the brain as a tractography. The analysis of tractographies has drawn attention from the machine learning and pattern recognition communities providing novel challenges such as finding an appropriate representation space for the data. Many of the current learning algorithms require the input to be from a vectorial space. This requirement contrasts with the intrinsic nature of the tractography because its basic elements, called streamlines or tracks, have different lengths and different number of points and for this reason they cannot be directly represented in a common vectorial space. In this work we propose the adoption of the dissimilarity representation which is an Euclidean embedding technique defined by selecting a set of streamlines called prototypes and then mapping any new streamline to the vector of distances from prototypes. We investigate the degree of approximation of this projection under different prototype selection policies and prototype set sizes in order to characterise its use on tractography data. Additionally we propose the use of a scalable approximation of the most effective prototype selection policy that provides fast and accurate dissimilarity approximations of complete tractographies.
MEG Decoding Across Subjects
Apr 16, 2014
Abstract:Brain decoding is a data analysis paradigm for neuroimaging experiments that is based on predicting the stimulus presented to the subject from the concurrent brain activity. In order to make inference at the group level, a straightforward but sometimes unsuccessful approach is to train a classifier on the trials of a group of subjects and then to test it on unseen trials from new subjects. The extreme difficulty is related to the structural and functional variability across the subjects. We call this approach "decoding across subjects". In this work, we address the problem of decoding across subjects for magnetoencephalographic (MEG) experiments and we provide the following contributions: first, we formally describe the problem and show that it belongs to a machine learning sub-field called transductive transfer learning (TTL). Second, we propose to use a simple TTL technique that accounts for the differences between train data and test data. Third, we propose the use of ensemble learning, and specifically of stacked generalization, to address the variability across subjects within train data, with the aim of producing more stable classifiers. On a face vs. scramble task MEG dataset of 16 subjects, we compare the standard approach of not modelling the differences across subjects, to the proposed one of combining TTL and ensemble learning. We show that the proposed approach is consistently more accurate than the standard one.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge