Laurent Petit
FIESTA: Autoencoders for accurate fiber segmentation in tractography
Dec 12, 2022Abstract:White matter bundle segmentation is a cornerstone of modern tractography to study the brain's structural connectivity in domains such as neurological disorders, neurosurgery, and aging. In this study, we present FIESTA (FIbEr Segmentation in Tractography using Autoencoders), a reliable and robust, fully automated, and easily semi-automatically calibrated pipeline based on deep autoencoders that can dissect and fully populate WM bundles. Our framework allows the transition from one anatomical bundle definition to another with marginal calibrating time. This pipeline is built upon FINTA, CINTA, and GESTA methods that demonstrated how autoencoders can be used successfully for streamline filtering, bundling, and streamline generation in tractography. Our proposed method improves bundling coverage by recovering hard-to-track bundles with generative sampling through the latent space seeding of the subject bundle and the atlas bundle. A latent space of streamlines is learned using autoencoder-based modeling combined with contrastive learning. Using an atlas of bundles in standard space (MNI), our proposed method segments new tractograms using the autoencoder latent distance between each tractogram streamline and its closest neighbor bundle in the atlas of bundles. Intra-subject bundle reliability is improved by recovering hard-to-track streamlines, using the autoencoder to generate new streamlines that increase each bundle's spatial coverage while remaining anatomically meaningful. Results show that our method is more reliable than state-of-the-art automated virtual dissection methods such as RecoBundles, RecoBundlesX, TractSeg, White Matter Analysis and XTRACT. Overall, these results show that our framework improves the practicality and usability of current state-of-the-art bundling framework
Supervised Tractogram Filtering using Geometric Deep Learning
Dec 06, 2022



Abstract:A tractogram is a virtual representation of the brain white matter. It is composed of millions of virtual fibers, encoded as 3D polylines, which approximate the white matter axonal pathways. To date, tractograms are the most accurate white matter representation and thus are used for tasks like presurgical planning and investigations of neuroplasticity, brain disorders, or brain networks. However, it is a well-known issue that a large portion of tractogram fibers is not anatomically plausible and can be considered artifacts of the tracking procedure. With Verifyber, we tackle the problem of filtering out such non-plausible fibers using a novel fully-supervised learning approach. Differently from other approaches based on signal reconstruction and/or brain topology regularization, we guide our method with the existing anatomical knowledge of the white matter. Using tractograms annotated according to anatomical principles, we train our model, Verifyber, to classify fibers as either anatomically plausible or non-plausible. The proposed Verifyber model is an original Geometric Deep Learning method that can deal with variable size fibers, while being invariant to fiber orientation. Our model considers each fiber as a graph of points, and by learning features of the edges between consecutive points via the proposed sequence Edge Convolution, it can capture the underlying anatomical properties. The output filtering results highly accurate and robust across an extensive set of experiments, and fast; with a 12GB GPU, filtering a tractogram of 1M fibers requires less than a minute. Verifyber implementation and trained models are available at https://github.com/FBK-NILab/verifyber.
Generative sampling in tractography using autoencoders (GESTA)
Apr 22, 2022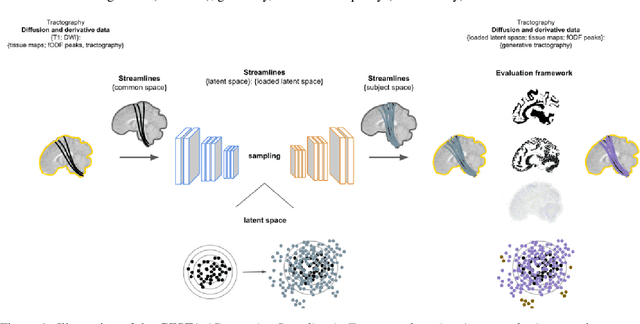
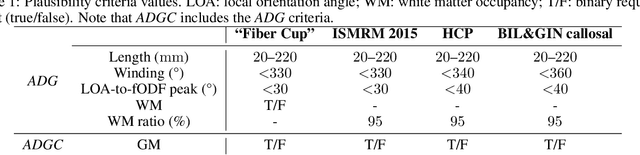
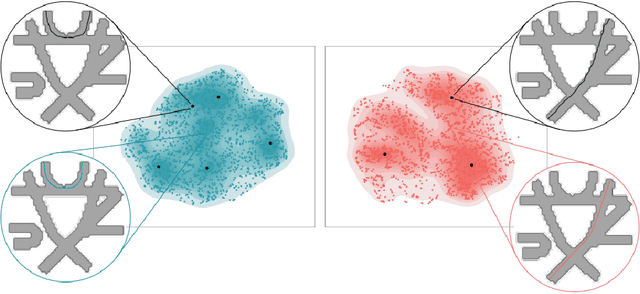
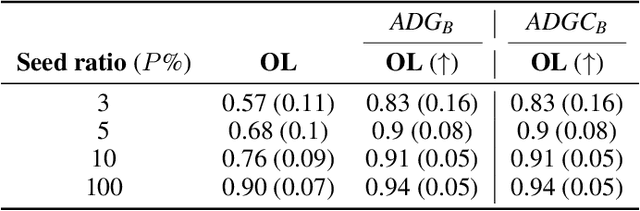
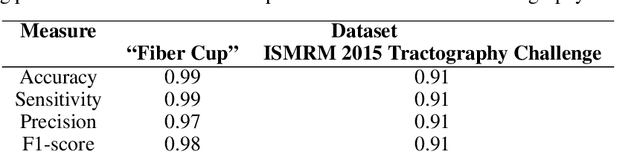
Abstract:Current tractography methods use the local orientation information to propagate streamlines from seed locations. Many such seeds provide streamlines that stop prematurely or fail to map the true pathways because some white matter bundles are "harder-to-track" than others. This results in tractography reconstructions with poor white and gray matter spatial coverage. In this work, we propose a generative, autoencoder-based method, named GESTA (Generative Sampling in Tractography using Autoencoders), that produces streamlines with better spatial coverage. Compared to other deep learning methods, our autoencoder-based framework is not constrained by any prior or a fixed set of bundles. GESTA produces new and complete streamlines for any white matter bundle. GESTA is shown to be effective on both synthetic and human brain in vivo data. Our streamline evaluation framework ensures that the streamlines produced by GESTA are anatomically plausible and fit well to the local diffusion signal. The streamline evaluation criteria assess anatomy (white matter coverage), local orientation alignment (direction), geometry features of streamlines, and optionally, gray matter connectivity. The GESTA framework offers considerable gains in bundle coverage using a reduced set of seeding streamlines with a 1.5x improvement for the "Fiber Cup", and 6x for the ISMRM 2015 Tractography Challenge datasets. Similarly, it provides a 4x white matter volume increase on the BIL&GIN callosal homotopic dataset. It also successfully generates new streamlines in poorly populated bundles, such as the fornix and other hard-to-track bundles, on in vivo data. GESTA is thus the first deep tractography generative method that can improve white matter reconstruction of hard-to-track bundles.
Tractography filtering using autoencoders
Oct 07, 2020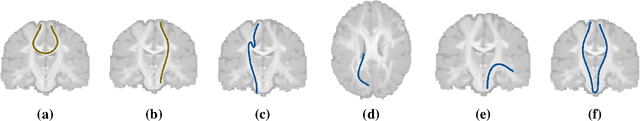
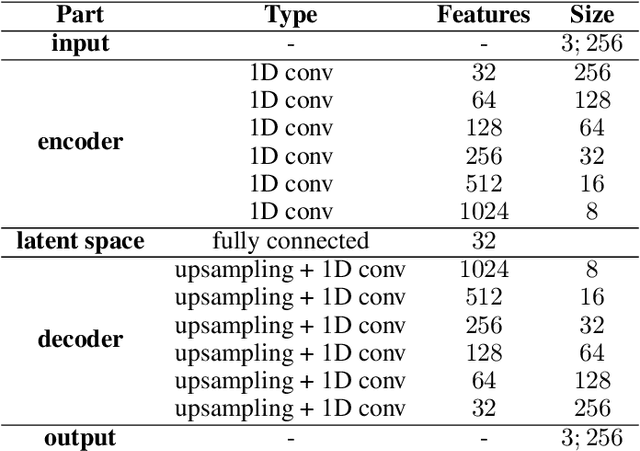
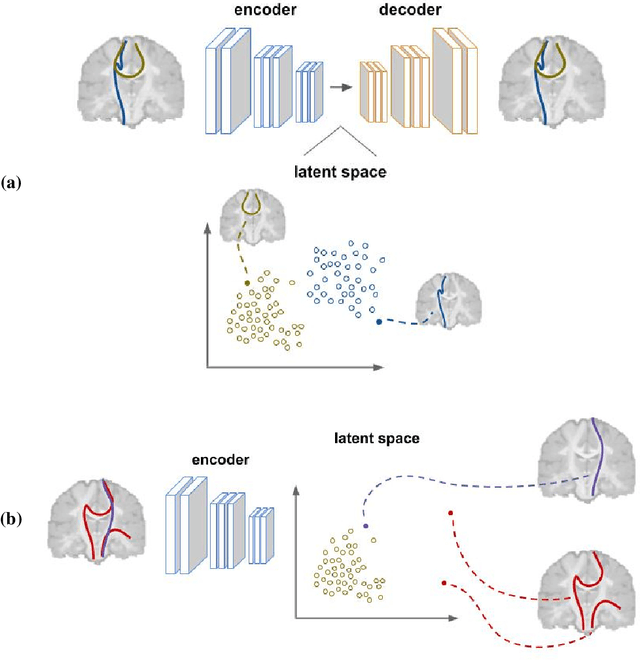
Abstract:Current brain white matter fiber tracking techniques show a number of problems, including: generating large proportions of streamlines that do not accurately describe the underlying anatomy; extracting streamlines that are not supported by the underlying diffusion signal; and under-representing some fiber populations, among others. In this paper, we describe a novel unsupervised learning method to filter streamlines from diffusion MRI tractography, and hence, to obtain more reliable tractograms. We show that a convolutional neural network autoencoder provides a straightforward and elegant way to learn a robust representation of brain streamlines, which can be used to filter undesired samples with a nearest neighbor algorithm. Our method, dubbed FINTA (Filtering in Tractography using Autoencoders) comes with several key advantages: training does not need labeled data, as it uses raw tractograms, it is fast and easily reproducible, it does not rely on the input diffusion MRI data, and thus, does not suffer from domain adaptation issues. We demonstrate the ability of FINTA to discriminate between "plausible" and "implausible" streamlines as well as to recover individual streamline group instances from a raw tractogram, from both synthetic and real human brain diffusion MRI tractography data, including partial tractograms. Results reveal that FINTA has a superior filtering performance compared to state-of-the-art methods. Together, this work brings forward a new deep learning framework in tractography based on autoencoders, and shows how it can be applied for filtering purposes. It sets the foundations for opening up new prospects towards more accurate and robust tractometry and connectivity diffusion MRI analyses, which may ultimately lead to improve the imaging of the white matter anatomy.
Tractogram filtering of anatomically non-plausible fibers with geometric deep learning
Mar 24, 2020



Abstract:Tractograms are virtual representations of the white matter fibers of the brain. They are of primary interest for tasks like presurgical planning, and investigation of neuroplasticity or brain disorders. Each tractogram is composed of millions of fibers encoded as 3D polylines. Unfortunately, a large portion of those fibers are not anatomically plausible and can be considered artifacts of the tracking algorithms. Common methods for tractogram filtering are based on signal reconstruction, a principled approach, but unable to consider the knowledge of brain anatomy. In this work, we address the problem of tractogram filtering as a supervised learning problem by exploiting the ground truth annotations obtained with a recent heuristic method, which labels fibers as either anatomically plausible or non-plausible according to well-established anatomical properties. The intuitive idea is to model a fiber as a point cloud and the goal is to investigate whether and how a geometric deep learning model might capture its anatomical properties. Our contribution is an extension of the Dynamic Edge Convolution model that exploits the sequential relations of points in a fiber and discriminates with high accuracy plausible/non-plausible fibers.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge