Philippe Poulin
FIESTA: Autoencoders for accurate fiber segmentation in tractography
Dec 12, 2022Abstract:White matter bundle segmentation is a cornerstone of modern tractography to study the brain's structural connectivity in domains such as neurological disorders, neurosurgery, and aging. In this study, we present FIESTA (FIbEr Segmentation in Tractography using Autoencoders), a reliable and robust, fully automated, and easily semi-automatically calibrated pipeline based on deep autoencoders that can dissect and fully populate WM bundles. Our framework allows the transition from one anatomical bundle definition to another with marginal calibrating time. This pipeline is built upon FINTA, CINTA, and GESTA methods that demonstrated how autoencoders can be used successfully for streamline filtering, bundling, and streamline generation in tractography. Our proposed method improves bundling coverage by recovering hard-to-track bundles with generative sampling through the latent space seeding of the subject bundle and the atlas bundle. A latent space of streamlines is learned using autoencoder-based modeling combined with contrastive learning. Using an atlas of bundles in standard space (MNI), our proposed method segments new tractograms using the autoencoder latent distance between each tractogram streamline and its closest neighbor bundle in the atlas of bundles. Intra-subject bundle reliability is improved by recovering hard-to-track streamlines, using the autoencoder to generate new streamlines that increase each bundle's spatial coverage while remaining anatomically meaningful. Results show that our method is more reliable than state-of-the-art automated virtual dissection methods such as RecoBundles, RecoBundlesX, TractSeg, White Matter Analysis and XTRACT. Overall, these results show that our framework improves the practicality and usability of current state-of-the-art bundling framework
Tractography and machine learning: Current state and open challenges
Feb 14, 2019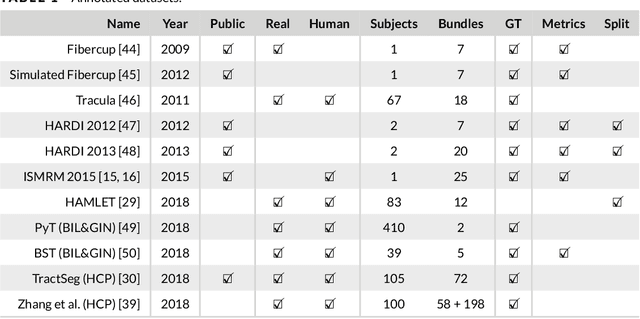
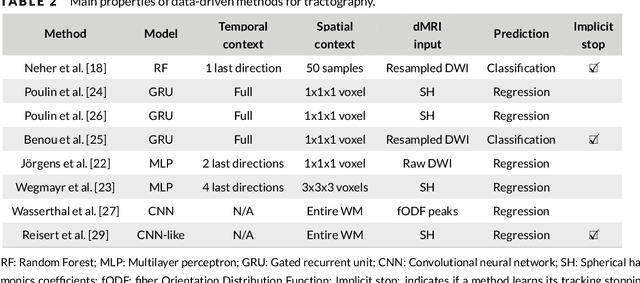
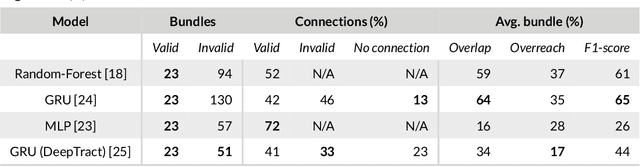
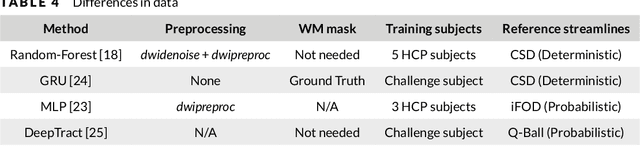
Abstract:Supervised machine learning (ML) algorithms have recently been proposed as an alternative to traditional tractography methods in order to address some of their weaknesses. They can be path-based and local-model-free, and easily incorporate anatomical priors to make contextual and non-local decisions that should help the tracking process. ML-based techniques have thus shown promising reconstructions of larger spatial extent of existing white matter bundles, promising reconstructions of less false positives, and promising robustness to known position and shape biases of current tractography techniques. But as of today, none of these ML-based methods have shown conclusive performances or have been adopted as a de facto solution to tractography. One reason for this might be the lack of well-defined and extensive frameworks to train, evaluate, and compare these methods. In this paper, we describe several datasets and evaluation tools that contain useful features for ML algorithms, along with the various methods proposed in the recent years. We then discuss the strategies that are used to evaluate and compare those methods, as well as their shortcomings. Finally, we describe the particular needs of ML tractography methods and discuss tangible solutions for future works.
Within-Brain Classification for Brain Tumor Segmentation
Oct 05, 2015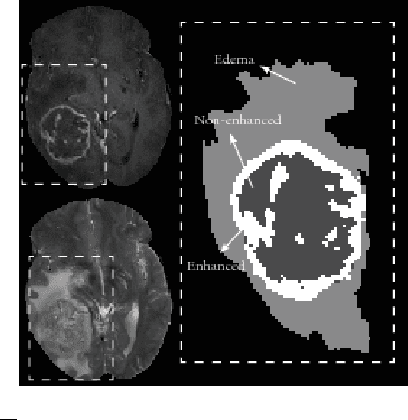
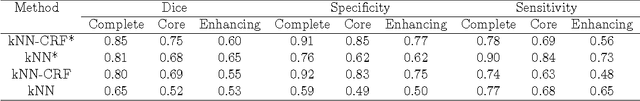
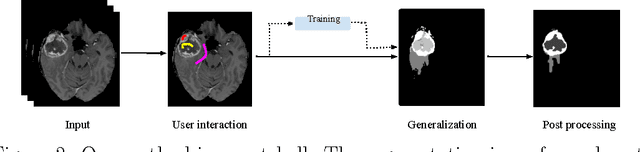
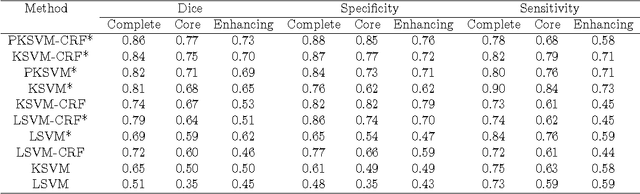
Abstract:Purpose: In this paper, we investigate a framework for interactive brain tumor segmentation which, at its core, treats the problem of interactive brain tumor segmentation as a machine learning problem. Methods: This method has an advantage over typical machine learning methods for this task where generalization is made across brains. The problem with these methods is that they need to deal with intensity bias correction and other MRI-specific noise. In this paper, we avoid these issues by approaching the problem as one of within brain generalization. Specifically, we propose a semi-automatic method that segments a brain tumor by training and generalizing within that brain only, based on some minimum user interaction. Conclusion: We investigate how adding spatial feature coordinates (i.e. $i$, $j$, $k$) to the intensity features can significantly improve the performance of different classification methods such as SVM, kNN and random forests. This would only be possible within an interactive framework. We also investigate the use of a more appropriate kernel and the adaptation of hyper-parameters specifically for each brain. Results: As a result of these experiments, we obtain an interactive method whose results reported on the MICCAI-BRATS 2013 dataset are the second most accurate compared to published methods, while using significantly less memory and processing power than most state-of-the-art methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge