François Rheault
Sensitivity of quantitative diffusion MRI tractography and microstructure to anisotropic spatial sampling
Sep 26, 2024Abstract:Purpose: Diffusion weighted MRI (dMRI) and its models of neural structure provide insight into human brain organization and variations in white matter. A recent study by McMaster, et al. showed that complex graph measures of the connectome, the graphical representation of a tractogram, vary with spatial sampling changes, but biases introduced by anisotropic voxels in the process have not been well characterized. This study uses microstructural measures (fractional anisotropy and mean diffusivity) and white matter bundle properties (bundle volume, length, and surface area) to further understand the effect of anisotropic voxels on microstructure and tractography. Methods: The statistical significance of the selected measures derived from dMRI data were assessed by comparing three white matter bundles at different spatial resolutions with 44 subjects from the Human Connectome Project Young Adult dataset scan/rescan data using the Wilcoxon Signed Rank test. The original isotropic resolution (1.25 mm isotropic) was explored with six anisotropic resolutions with 0.25 mm incremental steps in the z dimension. Then, all generated resolutions were upsampled to 1.25 mm isotropic and 1 mm isotropic. Results: There were statistically significant differences between at least one microstructural and one bundle measure at every resolution (p less than or equal to 0.05, corrected for multiple comparisons). Cohen's d coefficient evaluated the effect size of anisotropic voxels on microstructure and tractography. Conclusion: Fractional anisotropy and mean diffusivity cannot be recovered with basic up sampling from low quality data with gold standard data. However, the bundle measures from tractogram become more repeatable when voxels are resampled to 1 mm isotropic.
Harmonized connectome resampling for variance in voxel sizes
Aug 02, 2024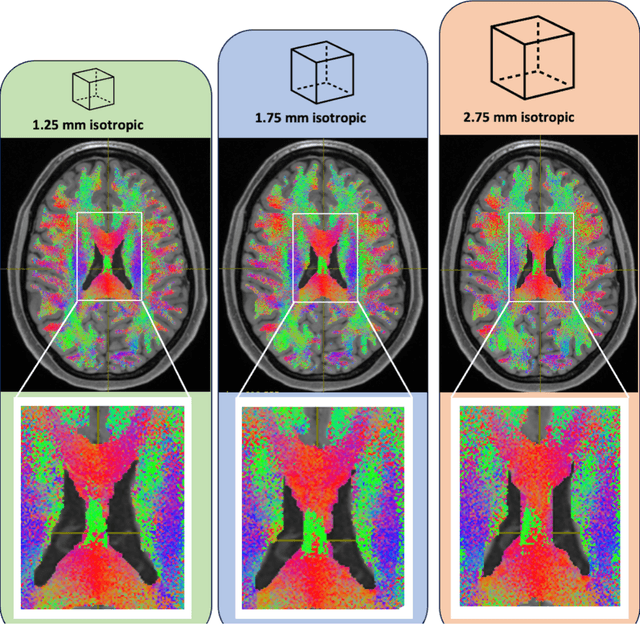
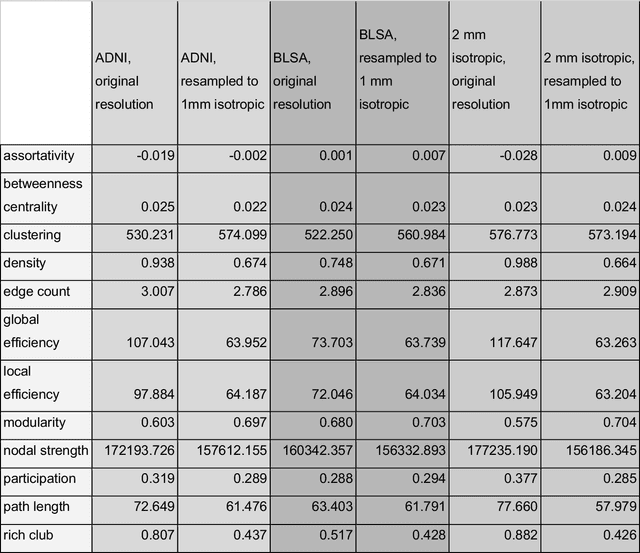
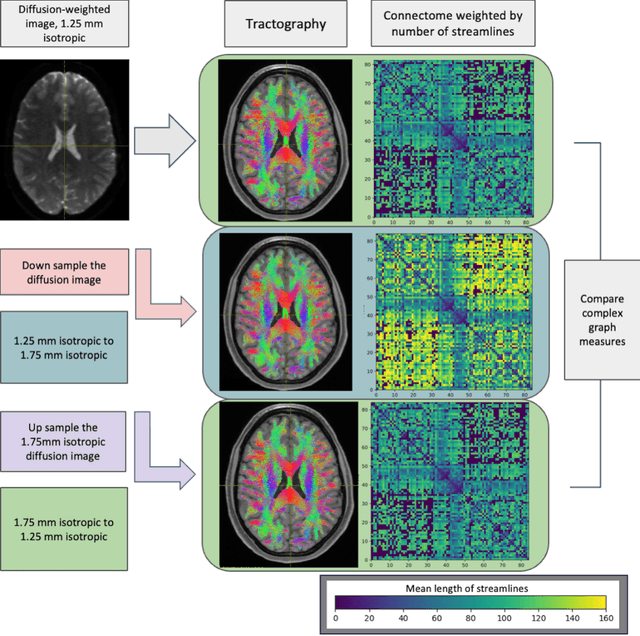
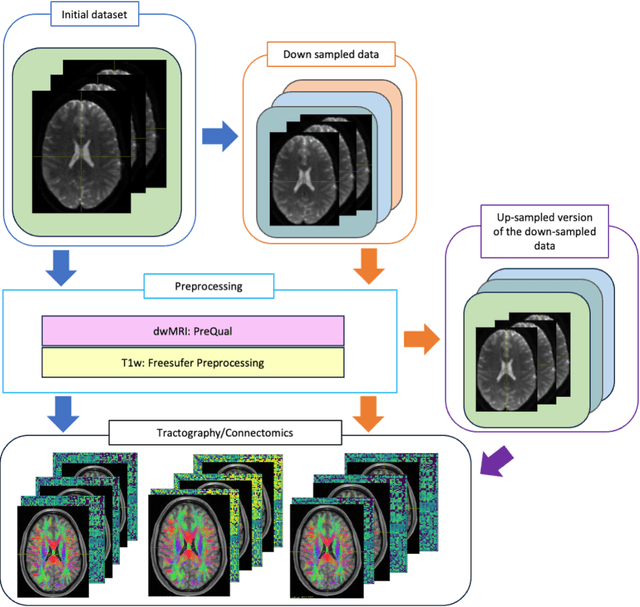
Abstract:To date, there has been no comprehensive study characterizing the effect of diffusion-weighted magnetic resonance imaging voxel resolution on the resulting connectome for high resolution subject data. Similarity in results improved with higher resolution, even after initial down-sampling. To ensure robust tractography and connectomes, resample data to 1 mm isotropic resolution.
FIESTA: Autoencoders for accurate fiber segmentation in tractography
Dec 12, 2022Abstract:White matter bundle segmentation is a cornerstone of modern tractography to study the brain's structural connectivity in domains such as neurological disorders, neurosurgery, and aging. In this study, we present FIESTA (FIbEr Segmentation in Tractography using Autoencoders), a reliable and robust, fully automated, and easily semi-automatically calibrated pipeline based on deep autoencoders that can dissect and fully populate WM bundles. Our framework allows the transition from one anatomical bundle definition to another with marginal calibrating time. This pipeline is built upon FINTA, CINTA, and GESTA methods that demonstrated how autoencoders can be used successfully for streamline filtering, bundling, and streamline generation in tractography. Our proposed method improves bundling coverage by recovering hard-to-track bundles with generative sampling through the latent space seeding of the subject bundle and the atlas bundle. A latent space of streamlines is learned using autoencoder-based modeling combined with contrastive learning. Using an atlas of bundles in standard space (MNI), our proposed method segments new tractograms using the autoencoder latent distance between each tractogram streamline and its closest neighbor bundle in the atlas of bundles. Intra-subject bundle reliability is improved by recovering hard-to-track streamlines, using the autoencoder to generate new streamlines that increase each bundle's spatial coverage while remaining anatomically meaningful. Results show that our method is more reliable than state-of-the-art automated virtual dissection methods such as RecoBundles, RecoBundlesX, TractSeg, White Matter Analysis and XTRACT. Overall, these results show that our framework improves the practicality and usability of current state-of-the-art bundling framework
Tractography filtering using autoencoders
Oct 07, 2020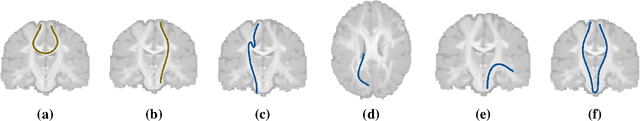
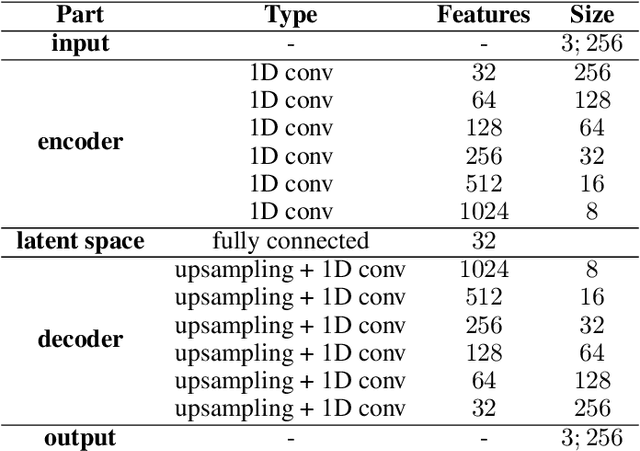
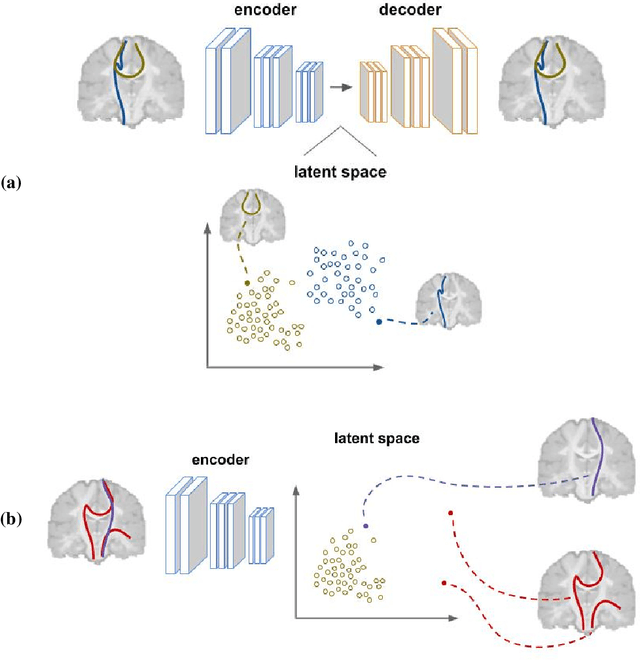
Abstract:Current brain white matter fiber tracking techniques show a number of problems, including: generating large proportions of streamlines that do not accurately describe the underlying anatomy; extracting streamlines that are not supported by the underlying diffusion signal; and under-representing some fiber populations, among others. In this paper, we describe a novel unsupervised learning method to filter streamlines from diffusion MRI tractography, and hence, to obtain more reliable tractograms. We show that a convolutional neural network autoencoder provides a straightforward and elegant way to learn a robust representation of brain streamlines, which can be used to filter undesired samples with a nearest neighbor algorithm. Our method, dubbed FINTA (Filtering in Tractography using Autoencoders) comes with several key advantages: training does not need labeled data, as it uses raw tractograms, it is fast and easily reproducible, it does not rely on the input diffusion MRI data, and thus, does not suffer from domain adaptation issues. We demonstrate the ability of FINTA to discriminate between "plausible" and "implausible" streamlines as well as to recover individual streamline group instances from a raw tractogram, from both synthetic and real human brain diffusion MRI tractography data, including partial tractograms. Results reveal that FINTA has a superior filtering performance compared to state-of-the-art methods. Together, this work brings forward a new deep learning framework in tractography based on autoencoders, and shows how it can be applied for filtering purposes. It sets the foundations for opening up new prospects towards more accurate and robust tractometry and connectivity diffusion MRI analyses, which may ultimately lead to improve the imaging of the white matter anatomy.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge