Carl Lemaire
FIESTA: Autoencoders for accurate fiber segmentation in tractography
Dec 12, 2022Abstract:White matter bundle segmentation is a cornerstone of modern tractography to study the brain's structural connectivity in domains such as neurological disorders, neurosurgery, and aging. In this study, we present FIESTA (FIbEr Segmentation in Tractography using Autoencoders), a reliable and robust, fully automated, and easily semi-automatically calibrated pipeline based on deep autoencoders that can dissect and fully populate WM bundles. Our framework allows the transition from one anatomical bundle definition to another with marginal calibrating time. This pipeline is built upon FINTA, CINTA, and GESTA methods that demonstrated how autoencoders can be used successfully for streamline filtering, bundling, and streamline generation in tractography. Our proposed method improves bundling coverage by recovering hard-to-track bundles with generative sampling through the latent space seeding of the subject bundle and the atlas bundle. A latent space of streamlines is learned using autoencoder-based modeling combined with contrastive learning. Using an atlas of bundles in standard space (MNI), our proposed method segments new tractograms using the autoencoder latent distance between each tractogram streamline and its closest neighbor bundle in the atlas of bundles. Intra-subject bundle reliability is improved by recovering hard-to-track streamlines, using the autoencoder to generate new streamlines that increase each bundle's spatial coverage while remaining anatomically meaningful. Results show that our method is more reliable than state-of-the-art automated virtual dissection methods such as RecoBundles, RecoBundlesX, TractSeg, White Matter Analysis and XTRACT. Overall, these results show that our framework improves the practicality and usability of current state-of-the-art bundling framework
Neural Teleportation
Dec 02, 2020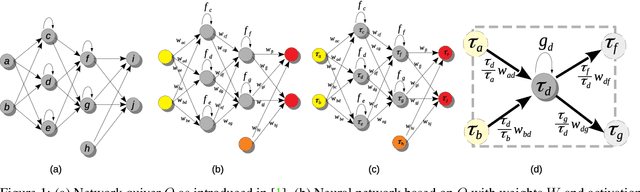
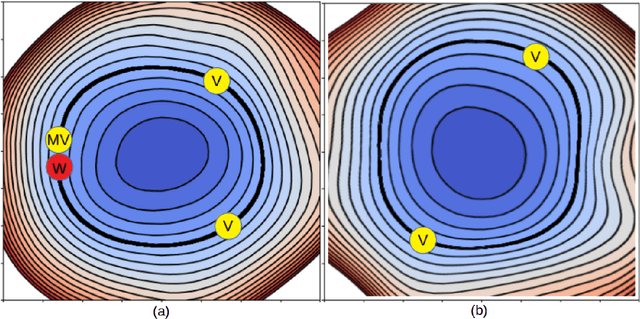
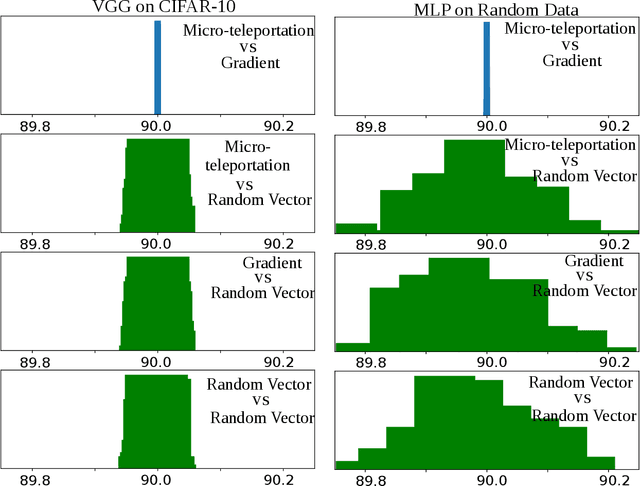
Abstract:In this paper, we explore a process called neural teleportation, a mathematical consequence of applying quiver representation theory to neural networks. Neural teleportation "teleports" a network to a new position in the weight space, while leaving its function unchanged. This concept generalizes the notion of positive scale invariance of ReLU networks to any network with any activation functions and any architecture. In this paper, we shed light on surprising and counter-intuitive consequences neural teleportation has on the loss landscape. In particular, we show that teleportation can be used to explore loss level curves, that it changes the loss landscape, sharpens global minima and boosts back-propagated gradients. From these observations, we demonstrate that teleportation accelerates training when used during initialization regardless of the model, its activation function, the loss function, and the training data. Our results can be reproduced with the code available here: https://github.com/vitalab/neuralteleportation.
Tractography filtering using autoencoders
Oct 07, 2020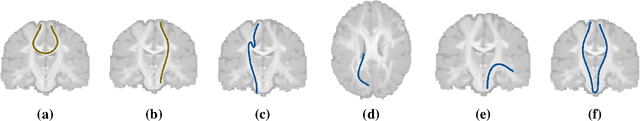
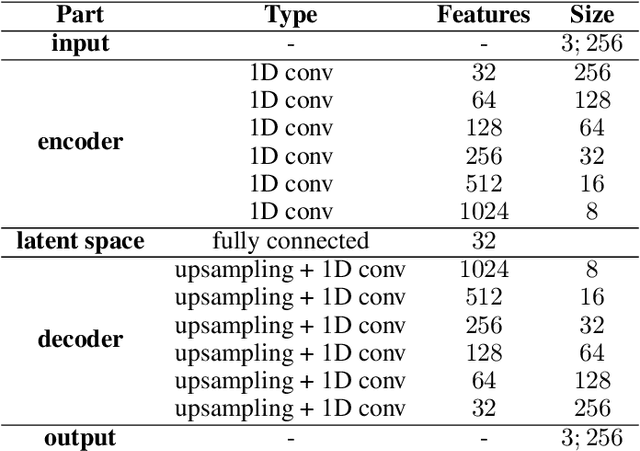
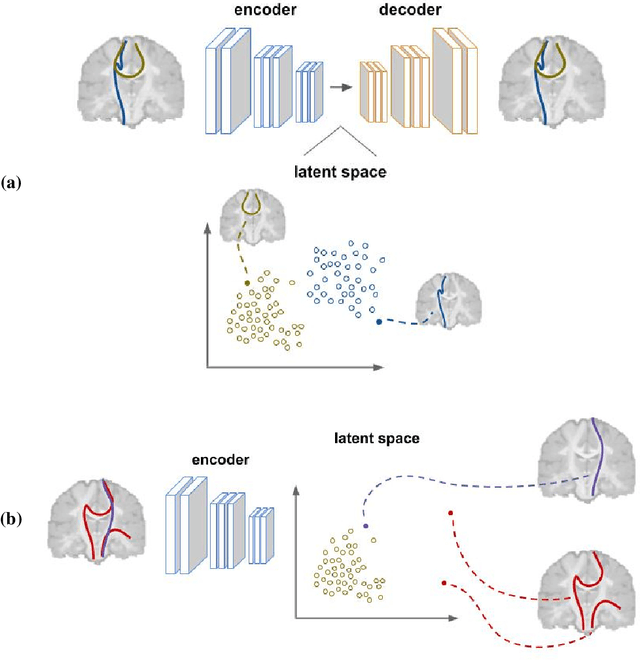
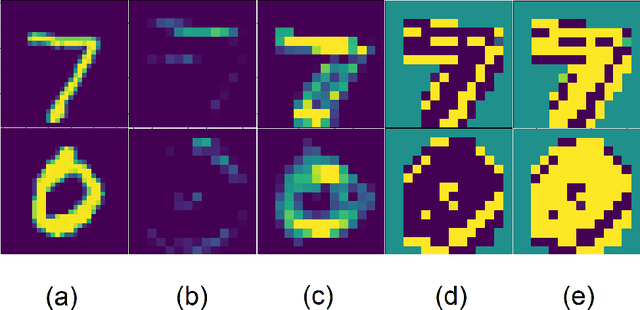
Abstract:Current brain white matter fiber tracking techniques show a number of problems, including: generating large proportions of streamlines that do not accurately describe the underlying anatomy; extracting streamlines that are not supported by the underlying diffusion signal; and under-representing some fiber populations, among others. In this paper, we describe a novel unsupervised learning method to filter streamlines from diffusion MRI tractography, and hence, to obtain more reliable tractograms. We show that a convolutional neural network autoencoder provides a straightforward and elegant way to learn a robust representation of brain streamlines, which can be used to filter undesired samples with a nearest neighbor algorithm. Our method, dubbed FINTA (Filtering in Tractography using Autoencoders) comes with several key advantages: training does not need labeled data, as it uses raw tractograms, it is fast and easily reproducible, it does not rely on the input diffusion MRI data, and thus, does not suffer from domain adaptation issues. We demonstrate the ability of FINTA to discriminate between "plausible" and "implausible" streamlines as well as to recover individual streamline group instances from a raw tractogram, from both synthetic and real human brain diffusion MRI tractography data, including partial tractograms. Results reveal that FINTA has a superior filtering performance compared to state-of-the-art methods. Together, this work brings forward a new deep learning framework in tractography based on autoencoders, and shows how it can be applied for filtering purposes. It sets the foundations for opening up new prospects towards more accurate and robust tractometry and connectivity diffusion MRI analyses, which may ultimately lead to improve the imaging of the white matter anatomy.
Structured Pruning of Neural Networks with Budget-Aware Regularization
Nov 23, 2018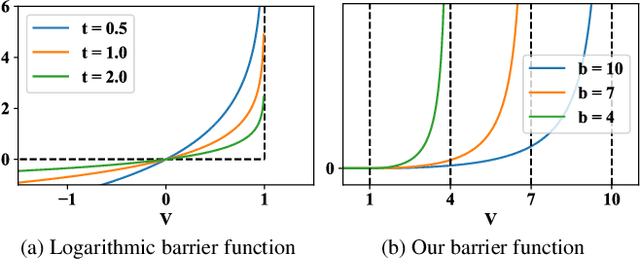


Abstract:Pruning methods have shown to be effective at reducing the size of deep neural networks while keeping accuracy almost intact. Among the most effective methods are those that prune a network while training it with a sparsity prior loss and learnable dropout parameters. A shortcoming of these approaches however is that neither the size nor the inference speed of the pruned network can be controlled directly; yet this is a key feature for targeting deployment of CNNs on low-power hardware. To overcome this, we introduce a budgeted regularized pruning framework for deep convolutional neural networks. Our approach naturally fits into traditional neural network training as it consists of a learnable masking layer, a novel budget-aware objective function, and the use of knowledge distillation. We also provide insights on how to prune a residual network and how this can lead to new architectures. Experimental results reveal that CNNs pruned with our method are more accurate and less compute-hungry than state-of-the-art methods. Also, our approach is more effective at preventing accuracy collapse in case of severe pruning; this allows us to attain pruning factors up to 16x without significantly affecting the accuracy.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge