Sandro Vega-Pons
Robustness of Generalized Median Computation for Consensus Learning in Arbitrary Spaces
Mar 07, 2025
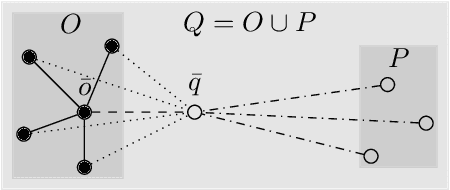

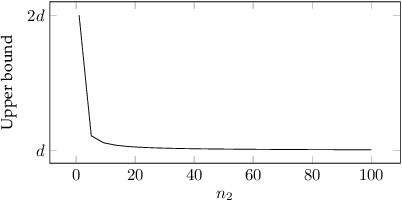
Abstract:Robustness in terms of outliers is an important topic and has been formally studied for a variety of problems in machine learning and computer vision. Generalized median computation is a special instance of consensus learning and a common approach to finding prototypes. Related research can be found in numerous problem domains with a broad range of applications. So far, however, robustness of generalized median has only been studied in a few specific spaces. To our knowledge, there is no robustness characterization in a general setting, i.e. for arbitrary spaces. We address this open issue in our work. The breakdown point >=0.5 is proved for generalized median with metric distance functions in general. We also study the detailed behavior in case of outliers from different perspectives. In addition, we present robustness results for weighted generalized median computation and non-metric distance functions. Given the importance of robustness, our work contributes to closing a gap in the literature. The presented results have general impact and applicability, e.g. providing deeper understanding of generalized median computation and practical guidance to avoid non-robust computation.
The Kernel Two-Sample Test for Brain Networks
Nov 19, 2015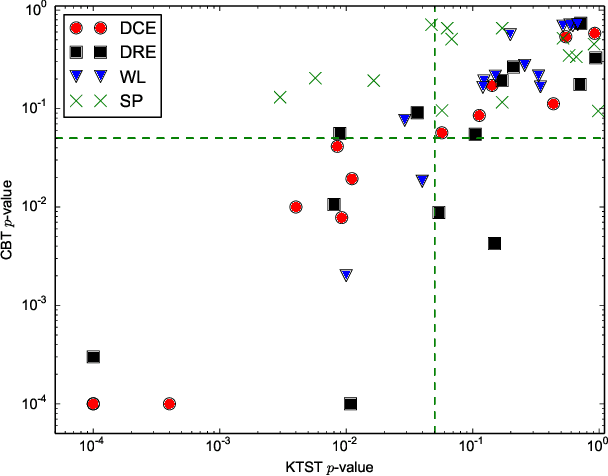



Abstract:In clinical and neuroscientific studies, systematic differences between two populations of brain networks are investigated in order to characterize mental diseases or processes. Those networks are usually represented as graphs built from neuroimaging data and studied by means of graph analysis methods. The typical machine learning approach to study these brain graphs creates a classifier and tests its ability to discriminate the two populations. In contrast to this approach, in this work we propose to directly test whether two populations of graphs are different or not, by using the kernel two-sample test (KTST), without creating the intermediate classifier. We claim that, in general, the two approaches provides similar results and that the KTST requires much less computation. Additionally, in the regime of low sample size, we claim that the KTST has lower frequency of Type II error than the classification approach. Besides providing algorithmic considerations to support these claims, we show strong evidence through experiments and one simulation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge