Elchanan Mossel
Learning and Testing Convex Functions
Nov 14, 2025Abstract:We consider the problems of \emph{learning} and \emph{testing} real-valued convex functions over Gaussian space. Despite the extensive study of function convexity across mathematics, statistics, and computer science, its learnability and testability have largely been examined only in discrete or restricted settings -- typically with respect to the Hamming distance, which is ill-suited for real-valued functions. In contrast, we study these problems in high dimensions under the standard Gaussian measure, assuming sample access to the function and a mild smoothness condition, namely Lipschitzness. A smoothness assumption is natural and, in fact, necessary even in one dimension: without it, convexity cannot be inferred from finitely many samples. As our main results, we give: - Learning Convex Functions: An agnostic proper learning algorithm for Lipschitz convex functions that achieves error $\varepsilon$ using $n^{O(1/\varepsilon^2)}$ samples, together with a complementary lower bound of $n^{\mathrm{poly}(1/\varepsilon)}$ samples in the \emph{correlational statistical query (CSQ)} model. - Testing Convex Functions: A tolerant (two-sided) tester for convexity of Lipschitz functions with the same sample complexity (as a corollary of our learning result), and a one-sided tester (which never rejects convex functions) using $O(\sqrt{n}/\varepsilon)^n$ samples.
Online Learning of Neural Networks
May 14, 2025



Abstract:We study online learning of feedforward neural networks with the sign activation function that implement functions from the unit ball in $\mathbb{R}^d$ to a finite label set $\{1, \ldots, Y\}$. First, we characterize a margin condition that is sufficient and in some cases necessary for online learnability of a neural network: Every neuron in the first hidden layer classifies all instances with some margin $\gamma$ bounded away from zero. Quantitatively, we prove that for any net, the optimal mistake bound is at most approximately $\mathtt{TS}(d,\gamma)$, which is the $(d,\gamma)$-totally-separable-packing number, a more restricted variation of the standard $(d,\gamma)$-packing number. We complement this result by constructing a net on which any learner makes $\mathtt{TS}(d,\gamma)$ many mistakes. We also give a quantitative lower bound of approximately $\mathtt{TS}(d,\gamma) \geq \max\{1/(\gamma \sqrt{d})^d, d\}$ when $\gamma \geq 1/2$, implying that for some nets and input sequences every learner will err for $\exp(d)$ many times, and that a dimension-free mistake bound is almost always impossible. To remedy this inevitable dependence on $d$, it is natural to seek additional natural restrictions to be placed on the network, so that the dependence on $d$ is removed. We study two such restrictions. The first is the multi-index model, in which the function computed by the net depends only on $k \ll d$ orthonormal directions. We prove a mistake bound of approximately $(1.5/\gamma)^{k + 2}$ in this model. The second is the extended margin assumption. In this setting, we assume that all neurons (in all layers) in the network classify every ingoing input from previous layer with margin $\gamma$ bounded away from zero. In this model, we prove a mistake bound of approximately $(\log Y)/ \gamma^{O(L)}$, where L is the depth of the network.
Low-dimensional Functions are Efficiently Learnable under Randomly Biased Distributions
Feb 10, 2025
Abstract:The problem of learning single index and multi index models has gained significant interest as a fundamental task in high-dimensional statistics. Many recent works have analysed gradient-based methods, particularly in the setting of isotropic data distributions, often in the context of neural network training. Such studies have uncovered precise characterisations of algorithmic sample complexity in terms of certain analytic properties of the target function, such as the leap, information, and generative exponents. These properties establish a quantitative separation between low and high complexity learning tasks. In this work, we show that high complexity cases are rare. Specifically, we prove that introducing a small random perturbation to the data distribution--via a random shift in the first moment--renders any Gaussian single index model as easy to learn as a linear function. We further extend this result to a class of multi index models, namely sparse Boolean functions, also known as Juntas.
Noise Sensitivity of Hierarchical Functions and Deep Learning Lower Bounds in General Product Measures
Feb 07, 2025Abstract:Recent works explore deep learning's success by examining functions or data with hierarchical structure. Complementarily, research on gradient descent performance for deep nets has shown that noise sensitivity of functions under independent and identically distributed (i.i.d.) Bernoulli inputs establishes learning complexity bounds. This paper aims to bridge these research streams by demonstrating that functions constructed through repeated composition of non-linear functions are noise sensitive under general product measures.
Efficiently Learning Markov Random Fields from Dynamics
Sep 09, 2024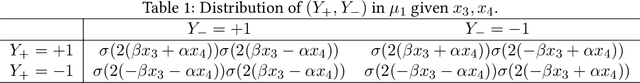
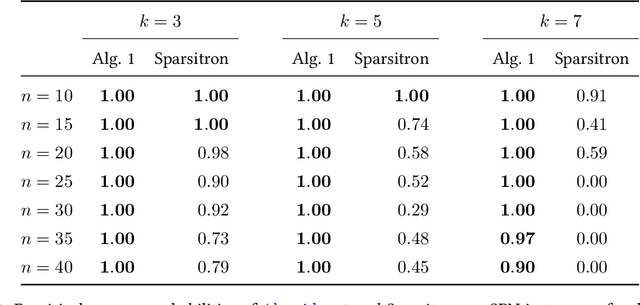
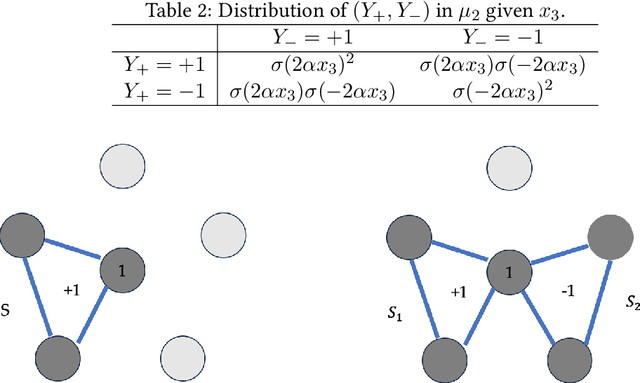
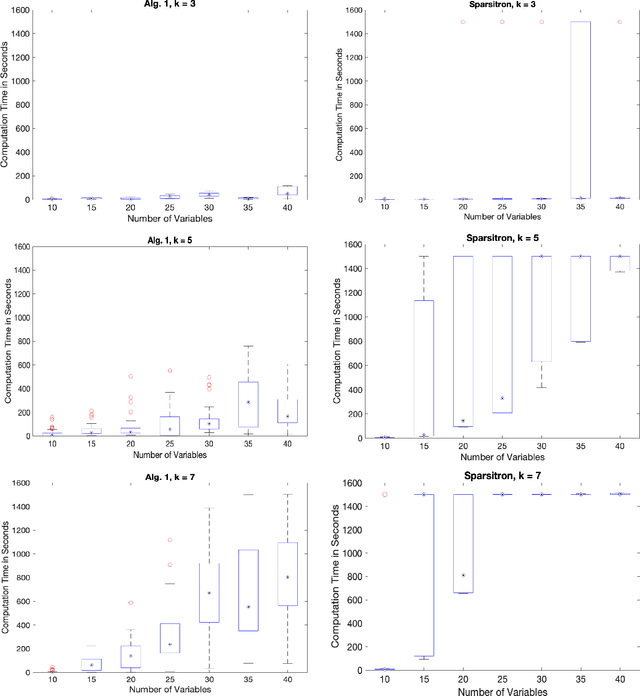
Abstract:An important task in high-dimensional statistics is learning the parameters or dependency structure of an undirected graphical model, or Markov random field (MRF). Much of the prior work on this problem assumes access to i.i.d. samples from the MRF distribution and state-of-the-art algorithms succeed using $n^{\Theta(k)}$ runtime, where $n$ is the dimension and $k$ is the order of the interactions. However, well-known reductions from the sparse parity with noise problem imply that given i.i.d. samples from a sparse, order-$k$ MRF, any learning algorithm likely requires $n^{\Omega(k)}$ time, impeding the potential for significant computational improvements. In this work, we demonstrate that these fundamental barriers for learning MRFs can surprisingly be completely circumvented when learning from natural, dynamical samples. We show that in bounded-degree MRFs, the dependency structure and parameters can be recovered using a trajectory of Glauber dynamics of length $O(n \log n)$ with runtime $O(n^2 \log n)$. The implicit constants depend only on the degree and non-degeneracy parameters of the model, but not the dimension $n$. In particular, learning MRFs from dynamics is $\textit{provably computationally easier}$ than learning from i.i.d. samples under standard hardness assumptions.
Sample-Efficient Linear Regression with Self-Selection Bias
Feb 22, 2024Abstract:We consider the problem of linear regression with self-selection bias in the unknown-index setting, as introduced in recent work by Cherapanamjeri, Daskalakis, Ilyas, and Zampetakis [STOC 2023]. In this model, one observes $m$ i.i.d. samples $(\mathbf{x}_{\ell},z_{\ell})_{\ell=1}^m$ where $z_{\ell}=\max_{i\in [k]}\{\mathbf{x}_{\ell}^T\mathbf{w}_i+\eta_{i,\ell}\}$, but the maximizing index $i_{\ell}$ is unobserved. Here, the $\mathbf{x}_{\ell}$ are assumed to be $\mathcal{N}(0,I_n)$ and the noise distribution $\mathbf{\eta}_{\ell}\sim \mathcal{D}$ is centered and independent of $\mathbf{x}_{\ell}$. We provide a novel and near optimally sample-efficient (in terms of $k$) algorithm to recover $\mathbf{w}_1,\ldots,\mathbf{w}_k\in \mathbb{R}^n$ up to additive $\ell_2$-error $\varepsilon$ with polynomial sample complexity $\tilde{O}(n)\cdot \mathsf{poly}(k,1/\varepsilon)$ and significantly improved time complexity $\mathsf{poly}(n,k,1/\varepsilon)+O(\log(k)/\varepsilon)^{O(k)}$. When $k=O(1)$, our algorithm runs in $\mathsf{poly}(n,1/\varepsilon)$ time, generalizing the polynomial guarantee of an explicit moment matching algorithm of Cherapanamjeri, et al. for $k=2$ and when it is known that $\mathcal{D}=\mathcal{N}(0,I_k)$. Our algorithm succeeds under significantly relaxed noise assumptions, and therefore also succeeds in the related setting of max-linear regression where the added noise is taken outside the maximum. For this problem, our algorithm is efficient in a much larger range of $k$ than the state-of-the-art due to Ghosh, Pananjady, Guntuboyina, and Ramchandran [IEEE Trans. Inf. Theory 2022] for not too small $\varepsilon$, and leads to improved algorithms for any $\varepsilon$ by providing a warm start for existing local convergence methods.
Reconstructing the Geometry of Random Geometric Graphs
Feb 14, 2024Abstract:Random geometric graphs are random graph models defined on metric spaces. Such a model is defined by first sampling points from a metric space and then connecting each pair of sampled points with probability that depends on their distance, independently among pairs. In this work, we show how to efficiently reconstruct the geometry of the underlying space from the sampled graph under the manifold assumption, i.e., assuming that the underlying space is a low dimensional manifold and that the connection probability is a strictly decreasing function of the Euclidean distance between the points in a given embedding of the manifold in $\mathbb{R}^N$. Our work complements a large body of work on manifold learning, where the goal is to recover a manifold from sampled points sampled in the manifold along with their (approximate) distances.
A Unified Approach to Learning Ising Models: Beyond Independence and Bounded Width
Nov 15, 2023Abstract:We revisit the problem of efficiently learning the underlying parameters of Ising models from data. Current algorithmic approaches achieve essentially optimal sample complexity when given i.i.d. samples from the stationary measure and the underlying model satisfies "width" bounds on the total $\ell_1$ interaction involving each node. We show that a simple existing approach based on node-wise logistic regression provably succeeds at recovering the underlying model in several new settings where these assumptions are violated: (1) Given dynamically generated data from a wide variety of local Markov chains, like block or round-robin dynamics, logistic regression recovers the parameters with optimal sample complexity up to $\log\log n$ factors. This generalizes the specialized algorithm of Bresler, Gamarnik, and Shah [IEEE Trans. Inf. Theory'18] for structure recovery in bounded degree graphs from Glauber dynamics. (2) For the Sherrington-Kirkpatrick model of spin glasses, given $\mathsf{poly}(n)$ independent samples, logistic regression recovers the parameters in most of the known high-temperature regime via a simple reduction to weaker structural properties of the measure. This improves on recent work of Anari, Jain, Koehler, Pham, and Vuong [ArXiv'23] which gives distribution learning at higher temperature. (3) As a simple byproduct of our techniques, logistic regression achieves an exponential improvement in learning from samples in the M-regime of data considered by Dutt, Lokhov, Vuffray, and Misra [ICML'21] as well as novel guarantees for learning from the adversarial Glauber dynamics of Chin, Moitra, Mossel, and Sandon [ArXiv'23]. Our approach thus significantly generalizes the elegant analysis of Wu, Sanghavi, and Dimakis [Neurips'19] without any algorithmic modification.
Combinative Cumulative Knowledge Processes
Sep 11, 2023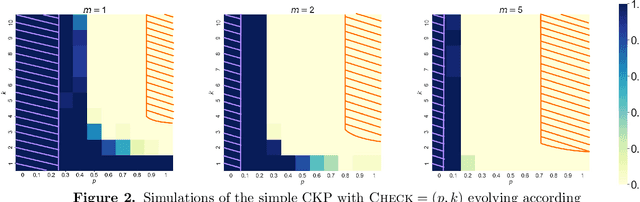
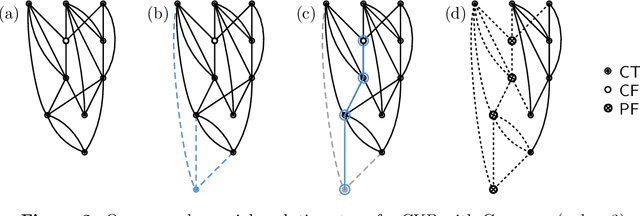
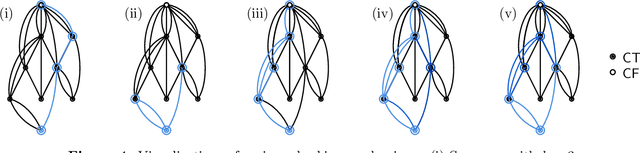
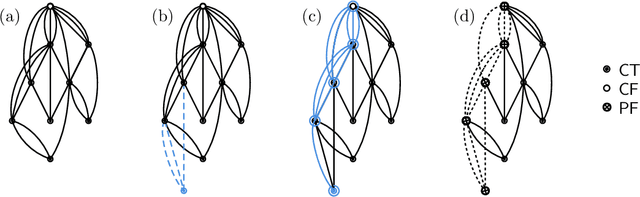
Abstract:We analyze Cumulative Knowledge Processes, introduced by Ben-Eliezer, Mikulincer, Mossel, and Sudan (ITCS 2023), in the setting of "directed acyclic graphs", i.e., when new units of knowledge may be derived by combining multiple previous units of knowledge. The main considerations in this model are the role of errors (when new units may be erroneous) and local checking (where a few antecedent units of knowledge are checked when a new unit of knowledge is discovered). The aforementioned work defined this model but only analyzed an idealized and simplified "tree-like" setting, i.e., a setting where new units of knowledge only depended directly on one previously generated unit of knowledge. The main goal of our work is to understand when the general process is safe, i.e., when the effect of errors remains under control. We provide some necessary and some sufficient conditions for safety. As in the earlier work, we demonstrate that the frequency of checking as well as the depth of the checks play a crucial role in determining safety. A key new parameter in the current work is the $\textit{combination factor}$ which is the distribution of the number of units $M$ of old knowledge that a new unit of knowledge depends on. Our results indicate that a large combination factor can compensate for a small depth of checking. The dependency of the safety on the combination factor is far from trivial. Indeed some of our main results are stated in terms of $\mathbb{E}\{1/M\}$ while others depend on $\mathbb{E}\{M\}$.
A Mathematical Model for Curriculum Learning
Jan 31, 2023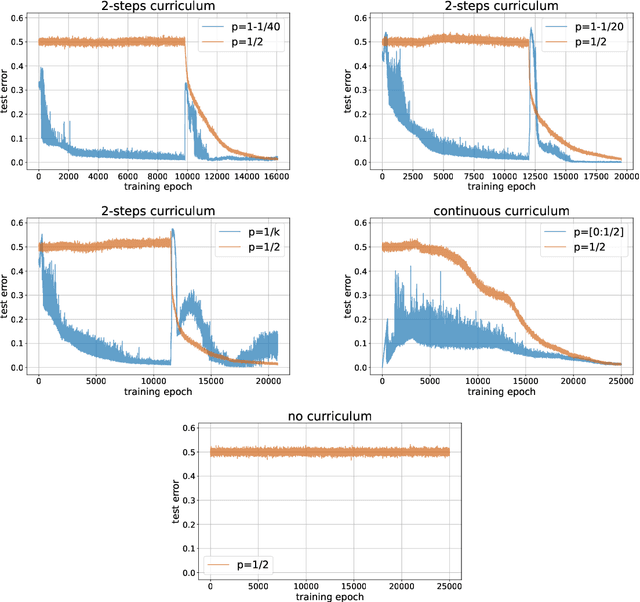
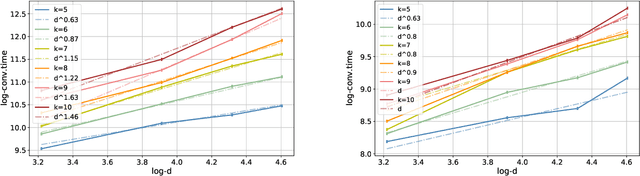
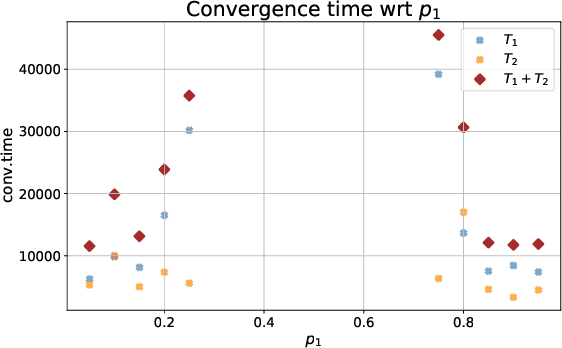
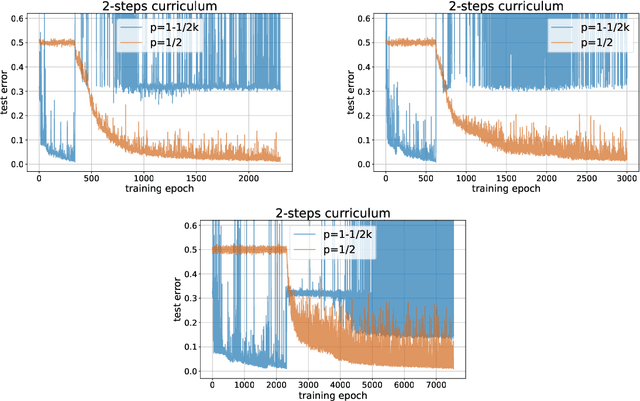
Abstract:Curriculum learning (CL) - training using samples that are generated and presented in a meaningful order - was introduced in the machine learning context around a decade ago. While CL has been extensively used and analysed empirically, there has been very little mathematical justification for its advantages. We introduce a CL model for learning the class of k-parities on d bits of a binary string with a neural network trained by stochastic gradient descent (SGD). We show that a wise choice of training examples, involving two or more product distributions, allows to reduce significantly the computational cost of learning this class of functions, compared to learning under the uniform distribution. We conduct experiments to support our analysis. Furthermore, we show that for another class of functions - namely the `Hamming mixtures' - CL strategies involving a bounded number of product distributions are not beneficial, while we conjecture that CL with unbounded many curriculum steps can learn this class efficiently.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge