Dou Xu
Asymmetric Co-Training with Explainable Cell Graph Ensembling for Histopathological Image Classification
Aug 24, 2023



Abstract:Convolutional neural networks excel in histopathological image classification, yet their pixel-level focus hampers explainability. Conversely, emerging graph convolutional networks spotlight cell-level features and medical implications. However, limited by their shallowness and suboptimal use of high-dimensional pixel data, GCNs underperform in multi-class histopathological image classification. To make full use of pixel-level and cell-level features dynamically, we propose an asymmetric co-training framework combining a deep graph convolutional network and a convolutional neural network for multi-class histopathological image classification. To improve the explainability of the entire framework by embedding morphological and topological distribution of cells, we build a 14-layer deep graph convolutional network to handle cell graph data. For the further utilization and dynamic interactions between pixel-level and cell-level information, we also design a co-training strategy to integrate the two asymmetric branches. Notably, we collect a private clinically acquired dataset termed LUAD7C, including seven subtypes of lung adenocarcinoma, which is rare and more challenging. We evaluated our approach on the private LUAD7C and public colorectal cancer datasets, showcasing its superior performance, explainability, and generalizability in multi-class histopathological image classification.
Graph Neural Networks for UnsupervisedDomain Adaptation of Histopathological ImageAnalytics
Aug 21, 2020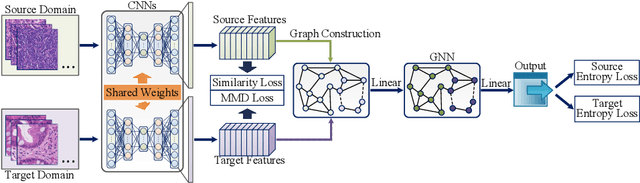
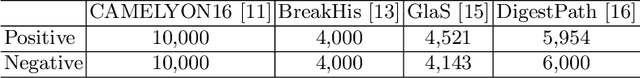
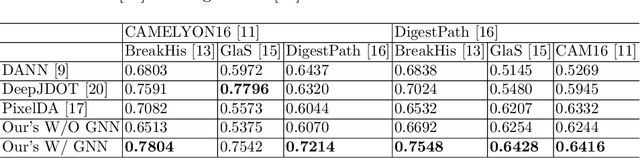
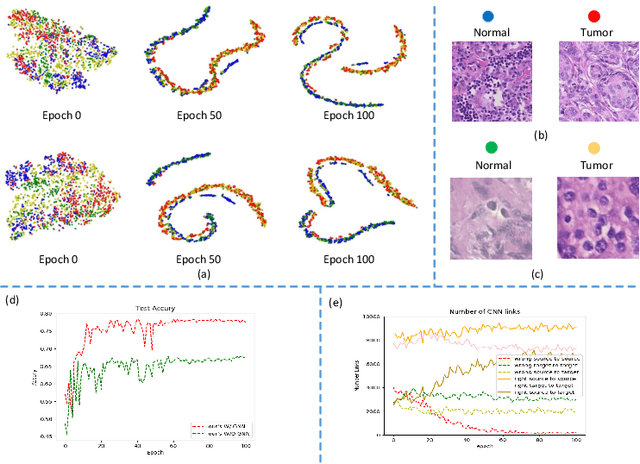
Abstract:Annotating histopathological images is a time-consuming andlabor-intensive process, which requires broad-certificated pathologistscarefully examining large-scale whole-slide images from cells to tissues.Recent frontiers of transfer learning techniques have been widely investi-gated for image understanding tasks with limited annotations. However,when applied for the analytics of histology images, few of them can effec-tively avoid the performance degradation caused by the domain discrep-ancy between the source training dataset and the target dataset, suchas different tissues, staining appearances, and imaging devices. To thisend, we present a novel method for the unsupervised domain adaptationin histopathological image analysis, based on a backbone for embeddinginput images into a feature space, and a graph neural layer for propa-gating the supervision signals of images with labels. The graph model isset up by connecting every image with its close neighbors in the embed-ded feature space. Then graph neural network is employed to synthesizenew feature representation from every image. During the training stage,target samples with confident inferences are dynamically allocated withpseudo labels. The cross-entropy loss function is used to constrain thepredictions of source samples with manually marked labels and targetsamples with pseudo labels. Furthermore, the maximum mean diversityis adopted to facilitate the extraction of domain-invariant feature repre-sentations, and contrastive learning is exploited to enhance the categorydiscrimination of learned features. In experiments of the unsupervised do-main adaptation for histopathological image classification, our methodachieves state-of-the-art performance on four public datasets
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge