Despina Kontos
Quantitative CT texture-based method to predict diagnosis and prognosis of fibrosing interstitial lung disease patterns
Jun 20, 2022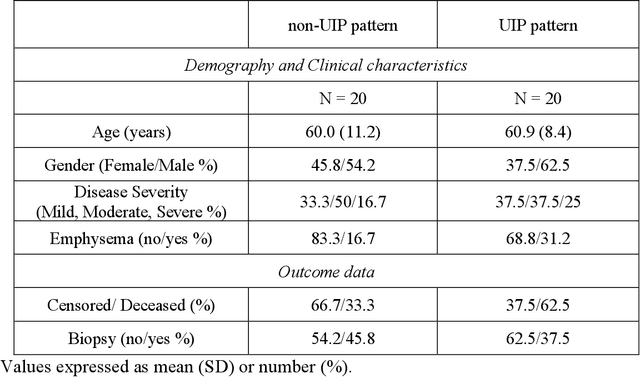
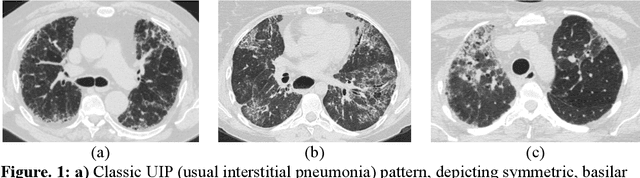
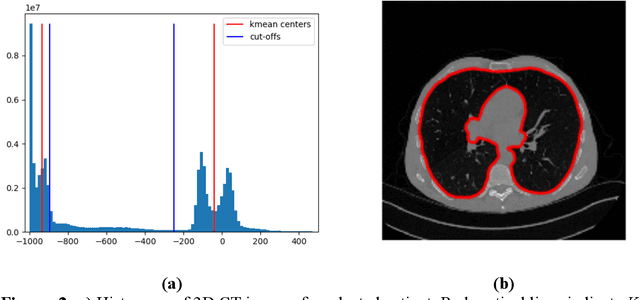
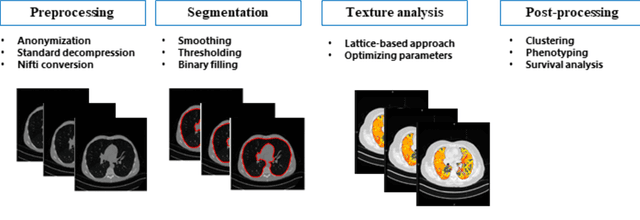
Abstract:Purpose: To utilize high-resolution quantitative CT (QCT) imaging features for prediction of diagnosis and prognosis in fibrosing interstitial lung diseases (ILD). Approach: 40 ILD patients (20 usual interstitial pneumonia (UIP), 20 non-UIP pattern ILD) were classified by expert consensus of 2 radiologists and followed for 7 years. Clinical variables were recorded. Following segmentation of the lung field, a total of 26 texture features were extracted using a lattice-based approach (TM model). The TM model was compared with previously histogram-based model (HM) for their abilities to classify UIP vs non-UIP. For prognostic assessment, survival analysis was performed comparing the expert diagnostic labels versus TM metrics. Results: In the classification analysis, the TM model outperformed the HM method with AUC of 0.70. While survival curves of UIP vs non-UIP expert labels in Cox regression analysis were not statistically different, TM QCT features allowed statistically significant partition of the cohort. Conclusions: TM model outperformed HM model in distinguishing UIP from non-UIP patterns. Most importantly, TM allows for partitioning of the cohort into distinct survival groups, whereas expert UIP vs non-UIP labeling does not. QCT TM models may improve diagnosis of ILD and offer more accurate prognostication, better guiding patient management.
MammoDL: Mammographic Breast Density Estimation using Federated Learning
Jun 11, 2022



Abstract:Assessing breast cancer risk from imaging remains a subjective process, in which radiologists employ computer aided detection (CAD) systems or qualitative visual assessment to estimate breast percent density (PD). More advanced machine learning (ML) models have become the most promising way to quantify breast cancer risk for early, accurate, and equitable diagnoses, but training such models in medical research is often restricted to small, single-institution data. Since patient demographics and imaging characteristics may vary considerably across imaging sites, models trained on single-institution data tend not to generalize well. In response to this problem, MammoDL is proposed, an open-source software tool that leverages UNet architecture to accurately estimate breast PD and complexity from digital mammography (DM). With the Open Federated Learning (OpenFL) library, this solution enables secure training on datasets across multiple institutions. MammoDL is a leaner, more flexible model than its predecessors, boasting improved generalization due to federation-enabled training on larger, more representative datasets.
GaNDLF: A Generally Nuanced Deep Learning Framework for Scalable End-to-End Clinical Workflows in Medical Imaging
Feb 26, 2021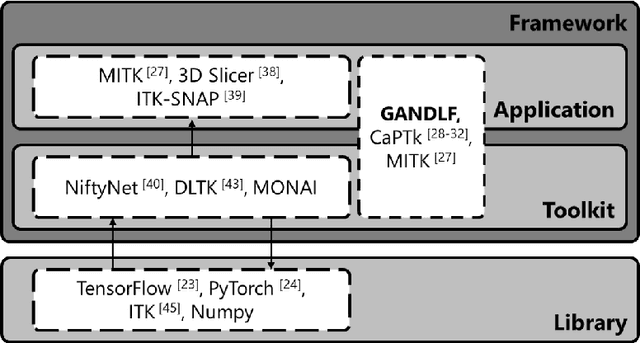
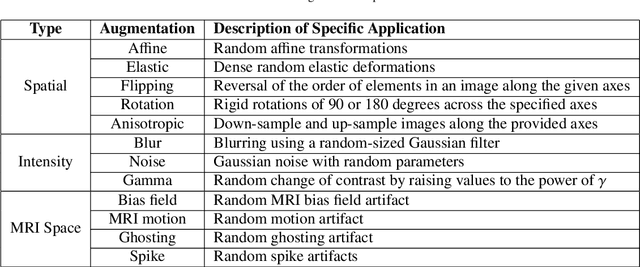
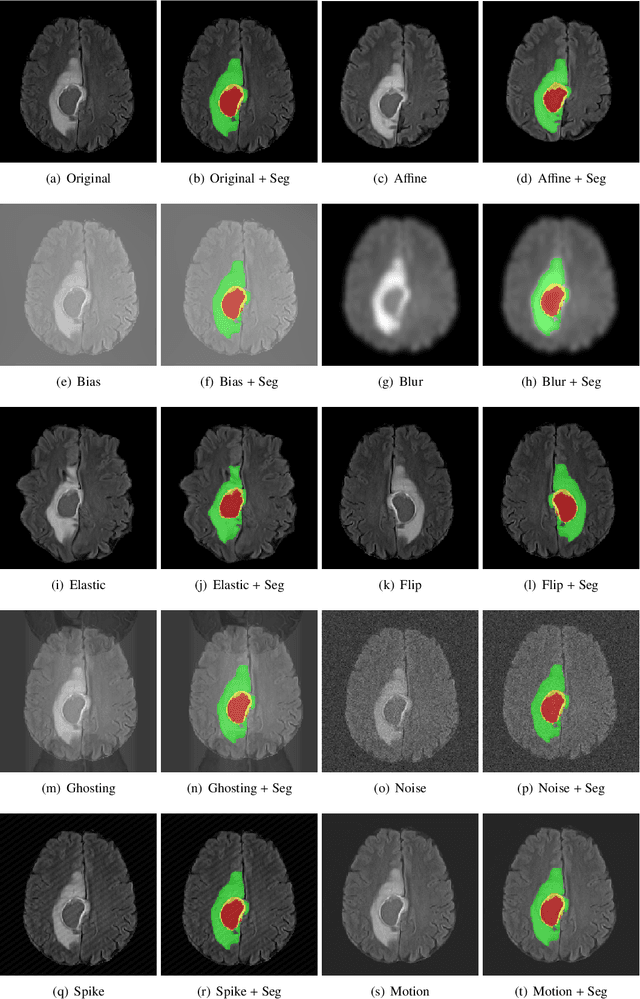
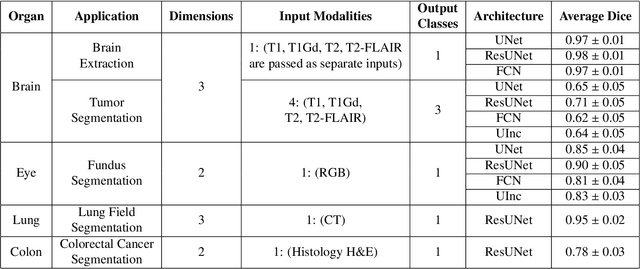
Abstract:Deep Learning (DL) has greatly highlighted the potential impact of optimized machine learning in both the scientific and clinical communities. The advent of open-source DL libraries from major industrial entities, such as TensorFlow (Google), PyTorch (Facebook), and MXNet (Apache), further contributes to DL promises on the democratization of computational analytics. However, increased technical and specialized background is required to develop DL algorithms, and the variability of implementation details hinders their reproducibility. Towards lowering the barrier and making the mechanism of DL development, training, and inference more stable, reproducible, and scalable, without requiring an extensive technical background, this manuscript proposes the \textbf{G}ener\textbf{a}lly \textbf{N}uanced \textbf{D}eep \textbf{L}earning \textbf{F}ramework (GaNDLF). With built-in support for $k$-fold cross-validation, data augmentation, multiple modalities and output classes, and multi-GPU training, as well as the ability to work with both radiographic and histologic imaging, GaNDLF aims to provide an end-to-end solution for all DL-related tasks, to tackle problems in medical imaging and provide a robust application framework for deployment in clinical workflows.
Deep-LIBRA: Artificial intelligence method for robust quantification of breast density with independent validation in breast cancer risk assessment
Nov 17, 2020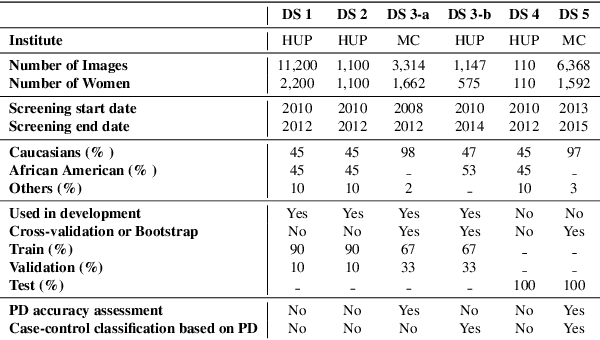
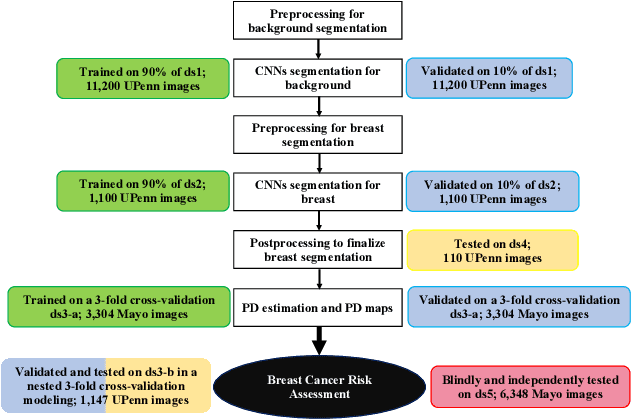
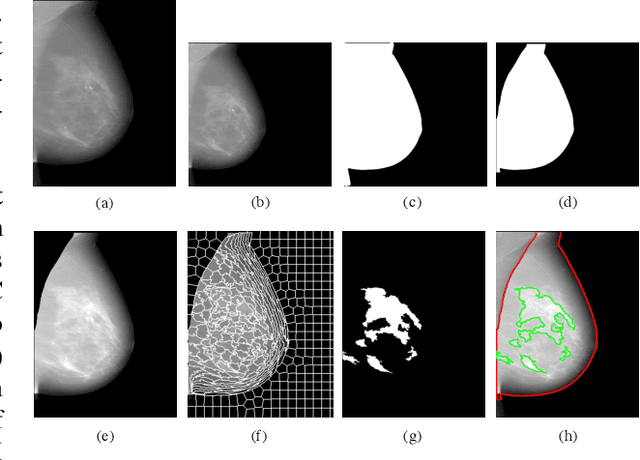

Abstract:Breast density is an important risk factor for breast cancer that also affects the specificity and sensitivity of screening mammography. Current federal legislation mandates reporting of breast density for all women undergoing breast screening. Clinically, breast density is assessed visually using the American College of Radiology Breast Imaging Reporting And Data System (BI-RADS) scale. Here, we introduce an artificial intelligence (AI) method to estimate breast percentage density (PD) from digital mammograms. Our method leverages deep learning (DL) using two convolutional neural network architectures to accurately segment the breast area. A machine-learning algorithm combining superpixel generation, texture feature analysis, and support vector machine is then applied to differentiate dense from non-dense tissue regions, from which PD is estimated. Our method has been trained and validated on a multi-ethnic, multi-institutional dataset of 15,661 images (4,437 women), and then tested on an independent dataset of 6,368 digital mammograms (1,702 women; cases=414) for both PD estimation and discrimination of breast cancer. On the independent dataset, PD estimates from Deep-LIBRA and an expert reader were strongly correlated (Spearman correlation coefficient = 0.90). Moreover, Deep-LIBRA yielded a higher breast cancer discrimination performance (area under the ROC curve, AUC = 0.611 [95% confidence interval (CI): 0.583, 0.639]) compared to four other widely-used research and commercial PD assessment methods (AUCs = 0.528 to 0.588). Our results suggest a strong agreement of PD estimates between Deep-LIBRA and gold-standard assessment by an expert reader, as well as improved performance in breast cancer risk assessment over state-of-the-art open-source and commercial methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge