Omid Haji Maghsoudi
Deep-LIBRA: Artificial intelligence method for robust quantification of breast density with independent validation in breast cancer risk assessment
Nov 17, 2020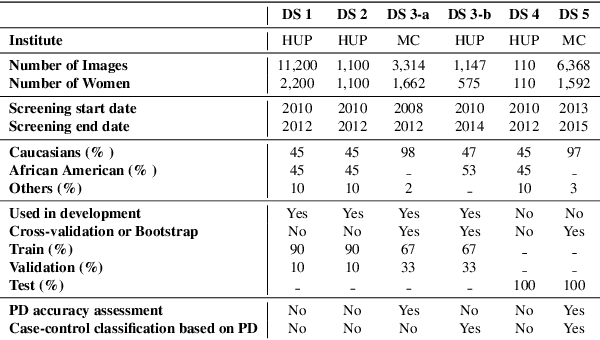
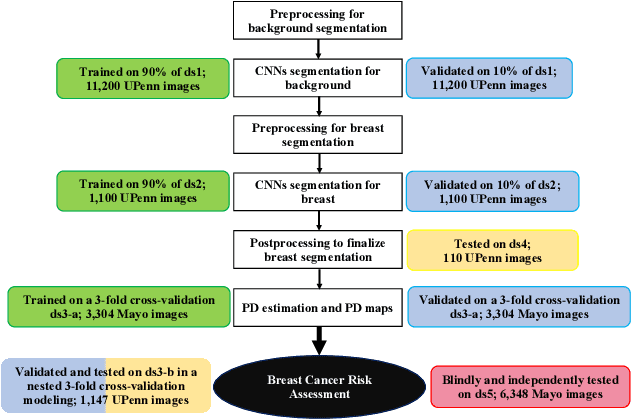
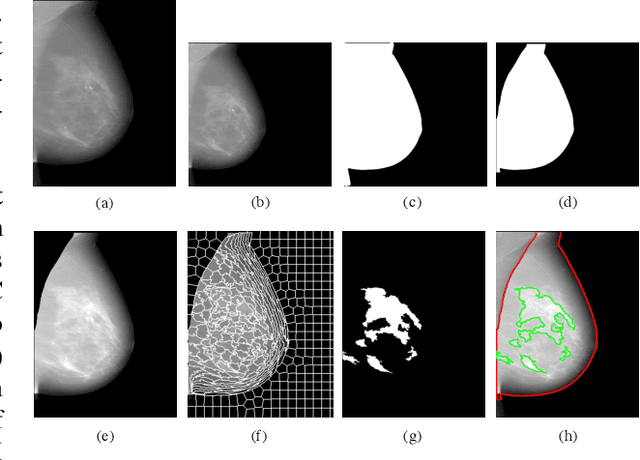

Abstract:Breast density is an important risk factor for breast cancer that also affects the specificity and sensitivity of screening mammography. Current federal legislation mandates reporting of breast density for all women undergoing breast screening. Clinically, breast density is assessed visually using the American College of Radiology Breast Imaging Reporting And Data System (BI-RADS) scale. Here, we introduce an artificial intelligence (AI) method to estimate breast percentage density (PD) from digital mammograms. Our method leverages deep learning (DL) using two convolutional neural network architectures to accurately segment the breast area. A machine-learning algorithm combining superpixel generation, texture feature analysis, and support vector machine is then applied to differentiate dense from non-dense tissue regions, from which PD is estimated. Our method has been trained and validated on a multi-ethnic, multi-institutional dataset of 15,661 images (4,437 women), and then tested on an independent dataset of 6,368 digital mammograms (1,702 women; cases=414) for both PD estimation and discrimination of breast cancer. On the independent dataset, PD estimates from Deep-LIBRA and an expert reader were strongly correlated (Spearman correlation coefficient = 0.90). Moreover, Deep-LIBRA yielded a higher breast cancer discrimination performance (area under the ROC curve, AUC = 0.611 [95% confidence interval (CI): 0.583, 0.639]) compared to four other widely-used research and commercial PD assessment methods (AUCs = 0.528 to 0.588). Our results suggest a strong agreement of PD estimates between Deep-LIBRA and gold-standard assessment by an expert reader, as well as improved performance in breast cancer risk assessment over state-of-the-art open-source and commercial methods.
Superpixel Based Segmentation and Classification of Polyps in Wireless Capsule Endoscopy
May 28, 2018



Abstract:Wireless Capsule Endoscopy (WCE) is a relatively new technology to record the entire GI trace, in vivo. The large amounts of frames captured during an examination cause difficulties for physicians to review all these frames. The need for reducing the reviewing time using some intelligent methods has been a challenge. Polyps are considered as growing tissues on the surface of intestinal tract not inside of an organ. Most polyps are not cancerous, but if one becomes larger than a centimeter, it can turn into cancer by great chance. The WCE frames provide the early stage possibility for detection of polyps. Here, the application of simple linear iterative clustering (SLIC) superpixel for segmentation of polyps in WCE frames is evaluated. Different SLIC superpixel numbers are examined to find the highest sensitivity for detection of polyps. The SLIC superpixel segmentation is promising to improve the results of previous studies. Finally, the superpixels were classified using a support vector machine (SVM) by extracting some texture and color features. The classification results showed a sensitivity of 91%.
Superpixels Based Marker Tracking Vs. Hue Thresholding In Rodent Biomechanics Application
May 28, 2018



Abstract:Examining locomotion has improved our basic understanding of motor control and aided in treating motor impairment. Mice and rats are premier models of human disease and increasingly the model systems of choice for basic neuroscience. High frame rates (250 Hz) are needed to quantify the kinematics of these running rodents. Manual tracking, especially for multiple markers, becomes time-consuming and impossible for large sample sizes. Therefore, the need for automatic segmentation of these markers has grown in recent years. We propose two methods to segment and track these markers: first, using SLIC superpixels segmentation with a tracker based on position, speed, shape, and color information of the segmented region in the previous frame; second, using a thresholding on hue channel following up with the same tracker. The comparison showed that the SLIC superpixels method was superior because the segmentation was more reliable and based on both color and spatial information.
Application of Superpixels to Segment Several Landmarks in Running Rodents
Apr 07, 2018



Abstract:Examining locomotion has improved our basic understanding of motor control and aided in treating motor impairment. Mice and rats are the model system of choice for basic neuroscience studies of human disease. High frame rates are needed to quantify the kinematics of running rodents, due to their high stride frequency. Manual tracking, especially for multiple body landmarks, becomes extremely time-consuming. To overcome these limitations, we proposed the use of superpixels based image segmentation as superpixels utilized both spatial and color information for segmentation. We segmented some parts of body and tested the success of segmentation as a function of color space and SLIC segment size. We used a simple merging function to connect the segmented regions considered as neighbor and having the same intensity value range. In addition, 28 features were extracted, and t-SNE was used to demonstrate how much the methods are capable to differentiate the regions. Finally, we compared the segmented regions to a manually outlined region. The results showed for segmentation, using the RGB image was slightly better compared to the hue channel. For merg- ing and classification, however, the hue representation was better as it captures the relevant color information in a single channel.
Feature Based Framework to Detect Diseases, Tumor, and Bleeding in Wireless Capsule Endoscopy
Jan 27, 2018



Abstract:Studying animal locomotion improves our understanding of motor control and aids in the treatment of motor impairment. Mice are a premier model of human disease and are the model system of choice for much of basic neuroscience. High frame rates (250 Hz) are needed to quantify the kinematics of these running rodents. Manual tracking, especially for multiple markers, becomes time-consuming and impossible. Therefore, an automated method is necessary. We propose a method to track the paws of the animal in the following manner: first, segmenting all the possible paws based on color; second, classifying the segmented objects using a support vector machine (SVM) and neural network (NN); third, classifying the objects using the kinematic features of the running animal, coupled with texture features from earlier frames; and finally, detecting and handling collisions to assure the correctness of labelled paws. The proposed method is validated in sixty 1,000 frame video sequences (4 seconds) captured by four cameras from five mice. The total sensitivity for tracking of the front and hind paw is 99.70% using the SVM classifier and 99.76% using the NN classifier. In addition, we show the feasibility of 3D reconstruction using the four camera system.
3D Based Landmark Tracker Using Superpixels Based Segmentation for Neuroscience and Biomechanics Studies
Nov 23, 2017



Abstract:Examining locomotion has improved our basic understanding of motor control and aided in treating motor impairment. Mice and rats are premier models of human disease and increasingly the model systems of choice for basic neuroscience. High frame rates (250 Hz) are needed to quantify the kinematics of these running rodents. Manual tracking, especially for multiple markers, becomes time-consuming and impossible for large sample sizes. Therefore, the need for automatic segmentation of these markers has grown in recent years. Here, we address this need by presenting a method to segment the markers using the SLIC superpixel method. The 2D coordinates on the image plane are projected to a 3D domain using direct linear transform (DLT) and a 3D Kalman filter has been used to predict the position of markers based on the speed and position of markers from the previous frames. Finally, a probabilistic function is used to find the best match among superpixels. The method is evaluated for different difficulties for tracking of the markers and it achieves 95% correct labeling of markers.
Superpixels Based Segmentation and SVM Based Classification Method to Distinguish Five Diseases from Normal Regions in Wireless Capsule Endoscopy
Nov 17, 2017



Abstract:Wireless Capsule Endoscopy (WCE) is relatively a new technology to examine the entire GI trace. During an examination, it captures more than 55,000 frames. Reviewing all these images is time-consuming and prone to human error. It has been a challenge to develop intelligent methods assisting physicians to review the frames. The WCE frames are captured in 8-bit color depths which provides enough a color range to detect abnormalities. Here, superpixel based methods are proposed to segment five diseases including: bleeding, Crohn's disease, Lymphangiectasia, Xanthoma, and Lymphoid hyperplasia. Two superpixels methods are compared to provide semantic segmentation of these prolific diseases: simple linear iterative clustering (SLIC) and quick shift (QS). The segmented superpixels were classified into two classes (normal and abnormal) by support vector machine (SVM) using texture and color features. For both superpixel methods, the accuracy, specificity, sensitivity, and precision (SLIC, QS) were around 92%, 93%, 93%, and 88%, respectively. However, SLIC was dramatically faster than QS.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge