Babak Haghighi
Quantitative CT texture-based method to predict diagnosis and prognosis of fibrosing interstitial lung disease patterns
Jun 20, 2022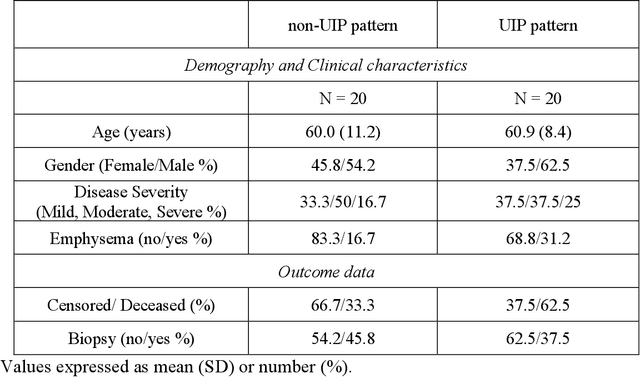
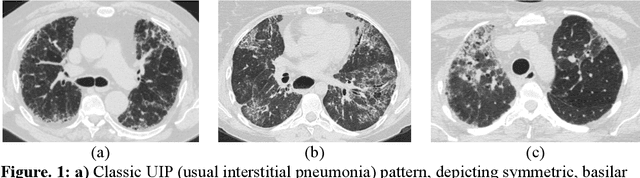
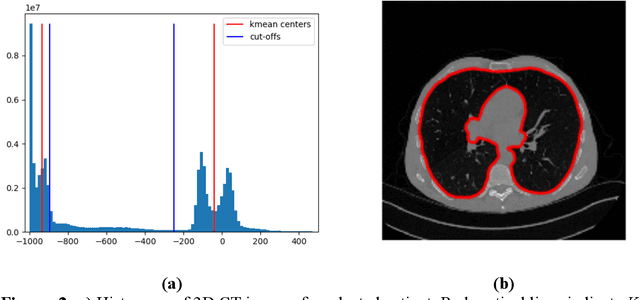
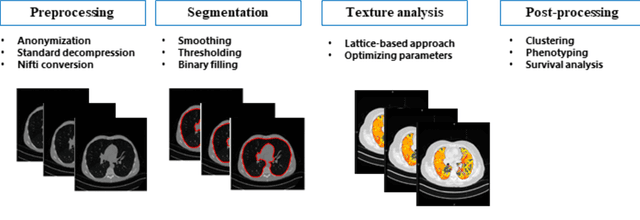
Abstract:Purpose: To utilize high-resolution quantitative CT (QCT) imaging features for prediction of diagnosis and prognosis in fibrosing interstitial lung diseases (ILD). Approach: 40 ILD patients (20 usual interstitial pneumonia (UIP), 20 non-UIP pattern ILD) were classified by expert consensus of 2 radiologists and followed for 7 years. Clinical variables were recorded. Following segmentation of the lung field, a total of 26 texture features were extracted using a lattice-based approach (TM model). The TM model was compared with previously histogram-based model (HM) for their abilities to classify UIP vs non-UIP. For prognostic assessment, survival analysis was performed comparing the expert diagnostic labels versus TM metrics. Results: In the classification analysis, the TM model outperformed the HM method with AUC of 0.70. While survival curves of UIP vs non-UIP expert labels in Cox regression analysis were not statistically different, TM QCT features allowed statistically significant partition of the cohort. Conclusions: TM model outperformed HM model in distinguishing UIP from non-UIP patterns. Most importantly, TM allows for partitioning of the cohort into distinct survival groups, whereas expert UIP vs non-UIP labeling does not. QCT TM models may improve diagnosis of ILD and offer more accurate prognostication, better guiding patient management.
GaNDLF: A Generally Nuanced Deep Learning Framework for Scalable End-to-End Clinical Workflows in Medical Imaging
Feb 26, 2021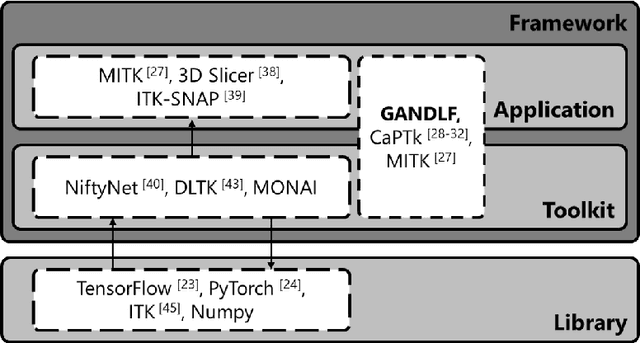
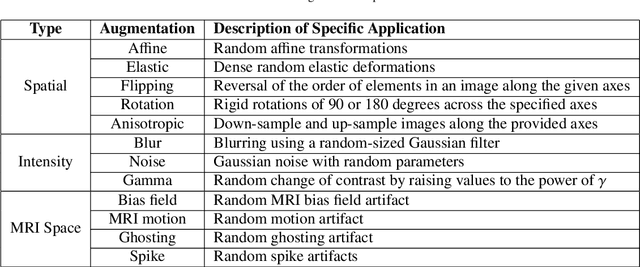
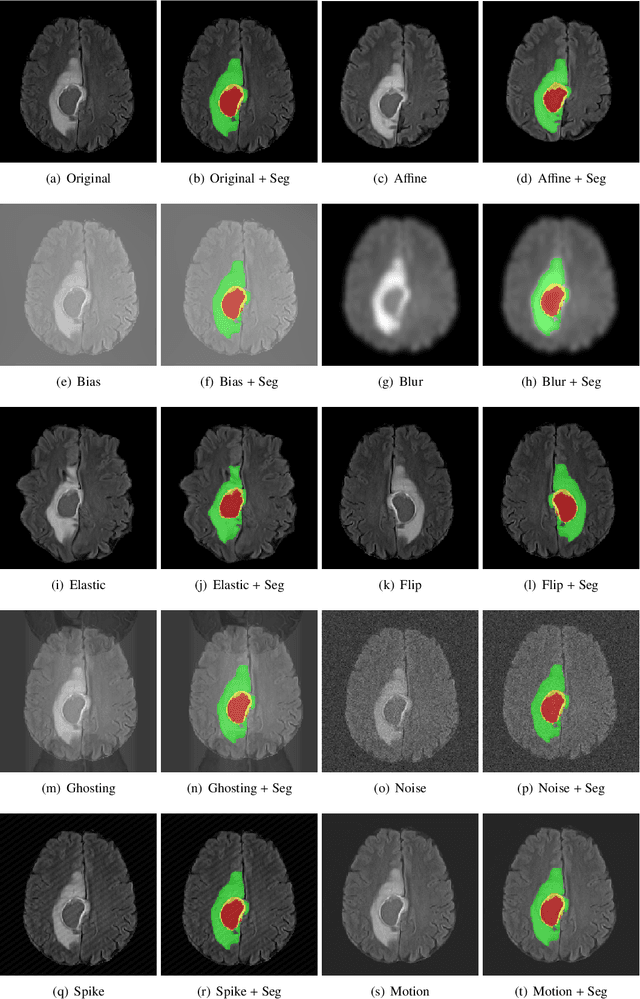
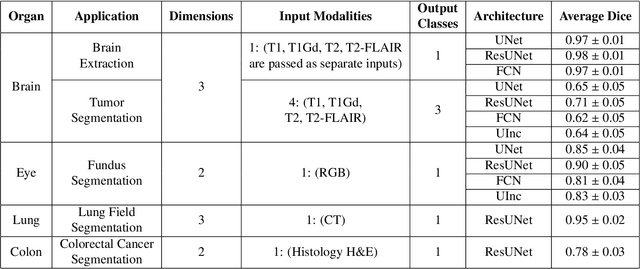
Abstract:Deep Learning (DL) has greatly highlighted the potential impact of optimized machine learning in both the scientific and clinical communities. The advent of open-source DL libraries from major industrial entities, such as TensorFlow (Google), PyTorch (Facebook), and MXNet (Apache), further contributes to DL promises on the democratization of computational analytics. However, increased technical and specialized background is required to develop DL algorithms, and the variability of implementation details hinders their reproducibility. Towards lowering the barrier and making the mechanism of DL development, training, and inference more stable, reproducible, and scalable, without requiring an extensive technical background, this manuscript proposes the \textbf{G}ener\textbf{a}lly \textbf{N}uanced \textbf{D}eep \textbf{L}earning \textbf{F}ramework (GaNDLF). With built-in support for $k$-fold cross-validation, data augmentation, multiple modalities and output classes, and multi-GPU training, as well as the ability to work with both radiographic and histologic imaging, GaNDLF aims to provide an end-to-end solution for all DL-related tasks, to tackle problems in medical imaging and provide a robust application framework for deployment in clinical workflows.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge