Davide Mottin
Automatic Annotation Augmentation Boosts Translation between Molecules and Natural Language
Feb 10, 2025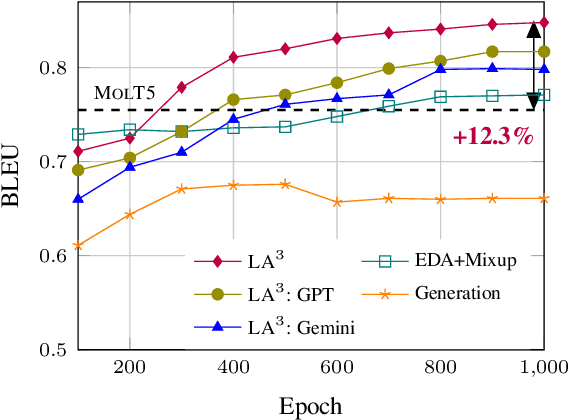
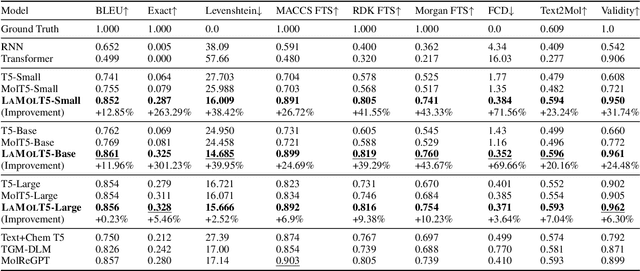
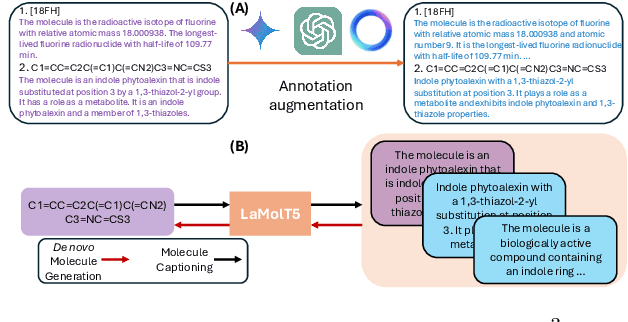
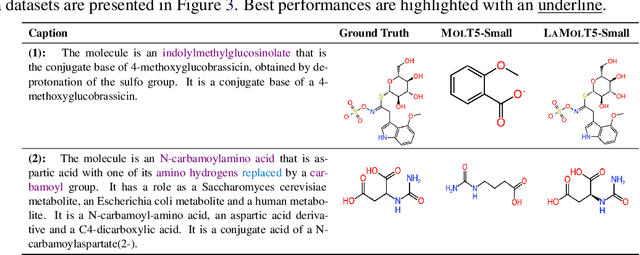
Abstract:Recent advancements in AI for biological research focus on integrating molecular data with natural language to accelerate drug discovery. However, the scarcity of high-quality annotations limits progress in this area. This paper introduces LA$^3$, a Language-based Automatic Annotation Augmentation framework that leverages large language models to augment existing datasets, thereby improving AI training. We demonstrate the effectiveness of LA$^3$ by creating an enhanced dataset, LaChEBI-20, where we systematically rewrite the annotations of molecules from an established dataset. These rewritten annotations preserve essential molecular information while providing more varied sentence structures and vocabulary. Using LaChEBI-20, we train LaMolT5 based on a benchmark architecture to learn the mapping between molecular representations and augmented annotations. Experimental results on text-based *de novo* molecule generation and molecule captioning demonstrate that LaMolT5 outperforms state-of-the-art models. Notably, incorporating LA$^3$ leads to improvements of up to 301% over the benchmark architecture. Furthermore, we validate the effectiveness of LA$^3$ notable applications in *image*, *text* and *graph* tasks, affirming its versatility and utility.
Exploring Graph Structure Comprehension Ability of Multimodal Large Language Models: Case Studies
Sep 13, 2024Abstract:Large Language Models (LLMs) have shown remarkable capabilities in processing various data structures, including graphs. While previous research has focused on developing textual encoding methods for graph representation, the emergence of multimodal LLMs presents a new frontier for graph comprehension. These advanced models, capable of processing both text and images, offer potential improvements in graph understanding by incorporating visual representations alongside traditional textual data. This study investigates the impact of graph visualisations on LLM performance across a range of benchmark tasks at node, edge, and graph levels. Our experiments compare the effectiveness of multimodal approaches against purely textual graph representations. The results provide valuable insights into both the potential and limitations of leveraging visual graph modalities to enhance LLMs' graph structure comprehension abilities.
An autoencoder for compressing angle-resolved photoemission spectroscopy data
Jul 05, 2024Abstract:Angle-resolved photoemission spectroscopy (ARPES) is a powerful experimental technique to determine the electronic structure of solids. Advances in light sources for ARPES experiments are currently leading to a vast increase of data acquisition rates and data quantity. On the other hand, access time to the most advanced ARPES instruments remains strictly limited, calling for fast, effective, and on-the-fly data analysis tools to exploit this time. In response to this need, we introduce ARPESNet, a versatile autoencoder network that efficiently summmarises and compresses ARPES datasets. We train ARPESNet on a large and varied dataset of 2-dimensional ARPES data extracted by cutting standard 3-dimensional ARPES datasets along random directions in $\mathbf{k}$. To test the data representation capacity of ARPESNet, we compare $k$-means clustering quality between data compressed by ARPESNet, data compressed by discrete cosine transform, and raw data, at different noise levels. ARPESNet data excels in clustering quality despite its high compression ratio.
Wiki Entity Summarization Benchmark
Jun 12, 2024



Abstract:Entity summarization aims to compute concise summaries for entities in knowledge graphs. Existing datasets and benchmarks are often limited to a few hundred entities and discard graph structure in source knowledge graphs. This limitation is particularly pronounced when it comes to ground-truth summaries, where there exist only a few labeled summaries for evaluation and training. We propose WikES, a comprehensive benchmark comprising of entities, their summaries, and their connections. Additionally, WikES features a dataset generator to test entity summarization algorithms in different areas of the knowledge graph. Importantly, our approach combines graph algorithms and NLP models as well as different data sources such that WikES does not require human annotation, rendering the approach cost-effective and generalizable to multiple domains. Finally, WikES is scalable and capable of capturing the complexities of knowledge graphs in terms of topology and semantics. WikES features existing datasets for comparison. Empirical studies of entity summarization methods confirm the usefulness of our benchmark. Data, code, and models are available at: https://github.com/msorkhpar/wiki-entity-summarization.
Benchmarking Large Language Models for Molecule Prediction Tasks
Mar 08, 2024



Abstract:Large Language Models (LLMs) stand at the forefront of a number of Natural Language Processing (NLP) tasks. Despite the widespread adoption of LLMs in NLP, much of their potential in broader fields remains largely unexplored, and significant limitations persist in their design and implementation. Notably, LLMs struggle with structured data, such as graphs, and often falter when tasked with answering domain-specific questions requiring deep expertise, such as those in biology and chemistry. In this paper, we explore a fundamental question: Can LLMs effectively handle molecule prediction tasks? Rather than pursuing top-tier performance, our goal is to assess how LLMs can contribute to diverse molecule tasks. We identify several classification and regression prediction tasks across six standard molecule datasets. Subsequently, we carefully design a set of prompts to query LLMs on these tasks and compare their performance with existing Machine Learning (ML) models, which include text-based models and those specifically designed for analysing the geometric structure of molecules. Our investigation reveals several key insights: Firstly, LLMs generally lag behind ML models in achieving competitive performance on molecule tasks, particularly when compared to models adept at capturing the geometric structure of molecules, highlighting the constrained ability of LLMs to comprehend graph data. Secondly, LLMs show promise in enhancing the performance of ML models when used collaboratively. Lastly, we engage in a discourse regarding the challenges and promising avenues to harness LLMs for molecule prediction tasks. The code and models are available at https://github.com/zhiqiangzhongddu/LLMaMol.
EvolMPNN: Predicting Mutational Effect on Homologous Proteins by Evolution Encoding
Feb 20, 2024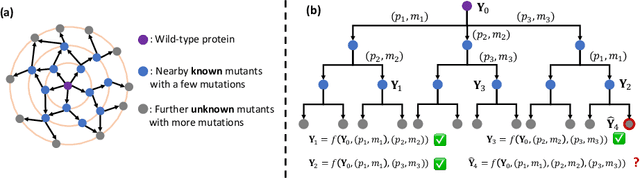
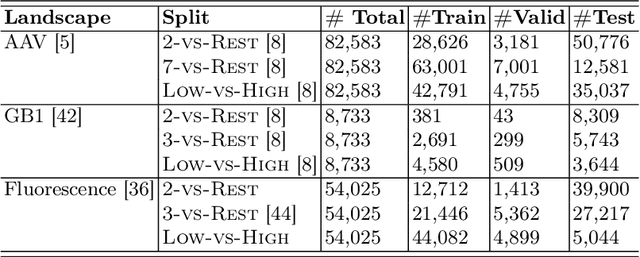
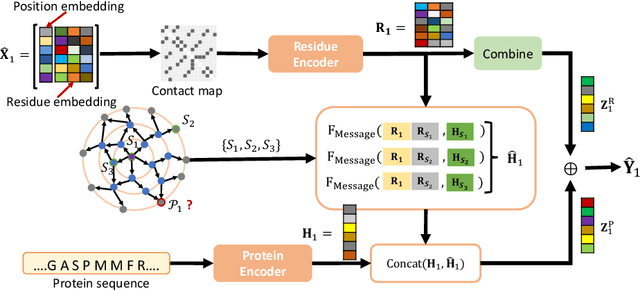
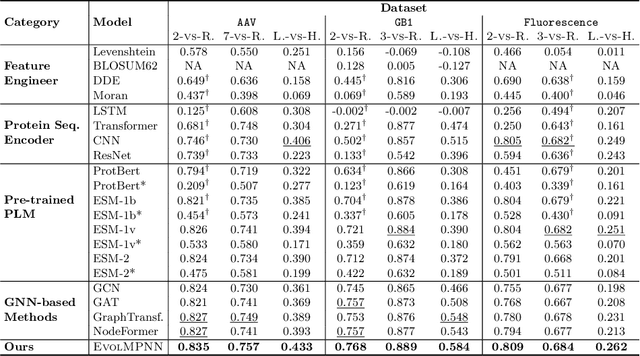
Abstract:Predicting protein properties is paramount for biological and medical advancements. Current protein engineering mutates on a typical protein, called the wild-type, to construct a family of homologous proteins and study their properties. Yet, existing methods easily neglect subtle mutations, failing to capture the effect on the protein properties. To this end, we propose EvolMPNN, Evolution-aware Message Passing Neural Network, to learn evolution-aware protein embeddings. EvolMPNN samples sets of anchor proteins, computes evolutionary information by means of residues and employs a differentiable evolution-aware aggregation scheme over these sampled anchors. This way EvolMPNNcan capture the mutation effect on proteins with respect to the anchor proteins. Afterwards, the aggregated evolution-aware embeddings are integrated with sequence embeddings to generate final comprehensive protein embeddings. Our model shows up to 6.4% better than state-of-the-art methods and attains 36x inference speedup in comparison with large pre-trained models.
Harnessing Large Language Models as Post-hoc Correctors
Feb 20, 2024Abstract:As Machine Learning (ML) models grow in size and demand higher-quality training data, the expenses associated with re-training and fine-tuning these models are escalating rapidly. Inspired by recent impressive achievements of Large Language Models (LLMs) in different fields, this paper delves into the question: can LLMs efficiently improve an ML's performance at a minimal cost? We show that, through our proposed training-free framework LlmCorr, an LLM can work as a post-hoc corrector to propose corrections for the predictions of an arbitrary ML model. In particular, we form a contextual knowledge database by incorporating the dataset's label information and the ML model's predictions on the validation dataset. Leveraging the in-context learning capability of LLMs, we ask the LLM to summarise the instances in which the ML model makes mistakes and the correlation between primary predictions and true labels. Following this, the LLM can transfer its acquired knowledge to suggest corrections for the ML model's predictions. Our experimental results on the challenging molecular predictions show that LlmCorr improves the performance of a number of models by up to 39%.
On the Robustness of Post-hoc GNN Explainers to Label Noise
Sep 08, 2023Abstract:Proposed as a solution to the inherent black-box limitations of graph neural networks (GNNs), post-hoc GNN explainers aim to provide precise and insightful explanations of the behaviours exhibited by trained GNNs. Despite their recent notable advancements in academic and industrial contexts, the robustness of post-hoc GNN explainers remains unexplored when confronted with label noise. To bridge this gap, we conduct a systematic empirical investigation to evaluate the efficacy of diverse post-hoc GNN explainers under varying degrees of label noise. Our results reveal several key insights: Firstly, post-hoc GNN explainers are susceptible to label perturbations. Secondly, even minor levels of label noise, inconsequential to GNN performance, harm the quality of generated explanations substantially. Lastly, we engage in a discourse regarding the progressive recovery of explanation effectiveness with escalating noise levels.
How Faithful are Self-Explainable GNNs?
Aug 29, 2023

Abstract:Self-explainable deep neural networks are a recent class of models that can output ante-hoc local explanations that are faithful to the model's reasoning, and as such represent a step forward toward filling the gap between expressiveness and interpretability. Self-explainable graph neural networks (GNNs) aim at achieving the same in the context of graph data. This begs the question: do these models fulfill their implicit guarantees in terms of faithfulness? In this extended abstract, we analyze the faithfulness of several self-explainable GNNs using different measures of faithfulness, identify several limitations -- both in the models themselves and in the evaluation metrics -- and outline possible ways forward.
ActUp: Analyzing and Consolidating tSNE and UMAP
May 12, 2023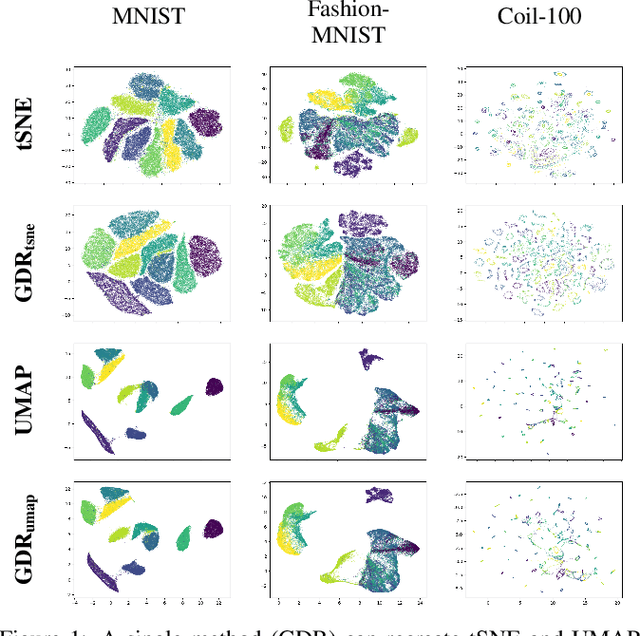

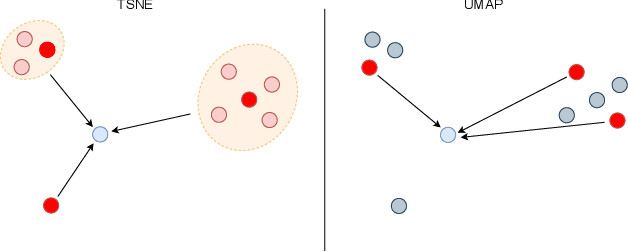
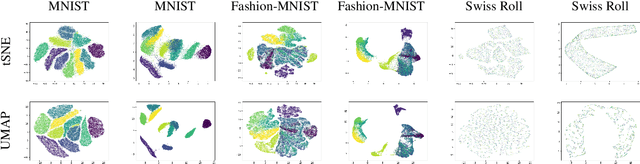
Abstract:tSNE and UMAP are popular dimensionality reduction algorithms due to their speed and interpretable low-dimensional embeddings. Despite their popularity, however, little work has been done to study their full span of differences. We theoretically and experimentally evaluate the space of parameters in both tSNE and UMAP and observe that a single one -- the normalization -- is responsible for switching between them. This, in turn, implies that a majority of the algorithmic differences can be toggled without affecting the embeddings. We discuss the implications this has on several theoretic claims behind UMAP, as well as how to reconcile them with existing tSNE interpretations. Based on our analysis, we provide a method (\ourmethod) that combines previously incompatible techniques from tSNE and UMAP and can replicate the results of either algorithm. This allows our method to incorporate further improvements, such as an acceleration that obtains either method's outputs faster than UMAP. We release improved versions of tSNE, UMAP, and \ourmethod that are fully plug-and-play with the traditional libraries at https://github.com/Andrew-Draganov/GiDR-DUN
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge