Kuangyu Zhou
Automatic Annotation Augmentation Boosts Translation between Molecules and Natural Language
Feb 10, 2025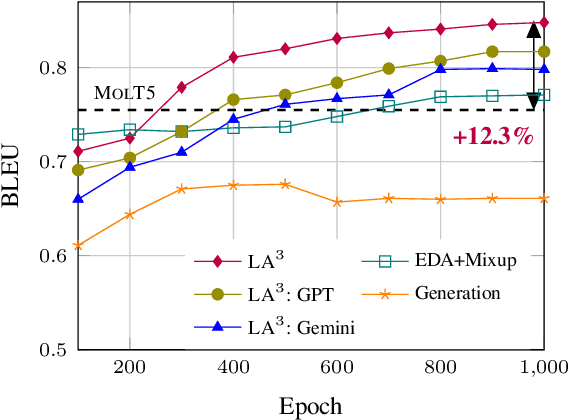
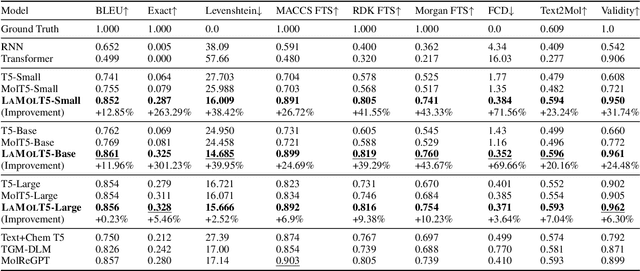
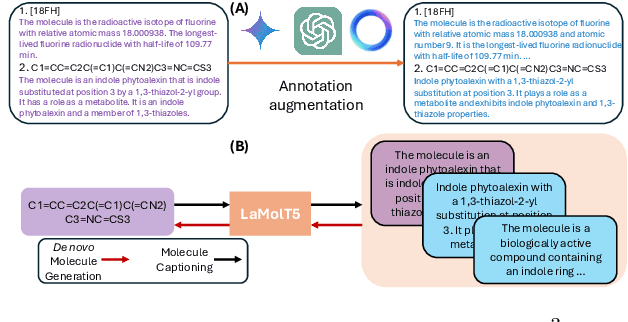
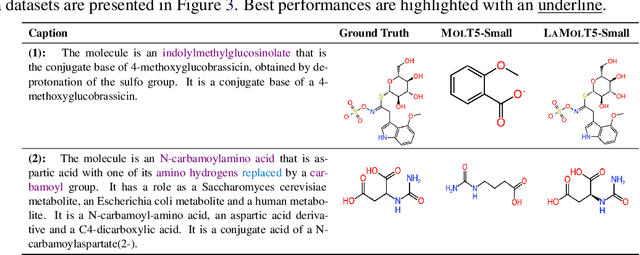
Abstract:Recent advancements in AI for biological research focus on integrating molecular data with natural language to accelerate drug discovery. However, the scarcity of high-quality annotations limits progress in this area. This paper introduces LA$^3$, a Language-based Automatic Annotation Augmentation framework that leverages large language models to augment existing datasets, thereby improving AI training. We demonstrate the effectiveness of LA$^3$ by creating an enhanced dataset, LaChEBI-20, where we systematically rewrite the annotations of molecules from an established dataset. These rewritten annotations preserve essential molecular information while providing more varied sentence structures and vocabulary. Using LaChEBI-20, we train LaMolT5 based on a benchmark architecture to learn the mapping between molecular representations and augmented annotations. Experimental results on text-based *de novo* molecule generation and molecule captioning demonstrate that LaMolT5 outperforms state-of-the-art models. Notably, incorporating LA$^3$ leads to improvements of up to 301% over the benchmark architecture. Furthermore, we validate the effectiveness of LA$^3$ notable applications in *image*, *text* and *graph* tasks, affirming its versatility and utility.
Benchmarking Large Language Models for Molecule Prediction Tasks
Mar 08, 2024



Abstract:Large Language Models (LLMs) stand at the forefront of a number of Natural Language Processing (NLP) tasks. Despite the widespread adoption of LLMs in NLP, much of their potential in broader fields remains largely unexplored, and significant limitations persist in their design and implementation. Notably, LLMs struggle with structured data, such as graphs, and often falter when tasked with answering domain-specific questions requiring deep expertise, such as those in biology and chemistry. In this paper, we explore a fundamental question: Can LLMs effectively handle molecule prediction tasks? Rather than pursuing top-tier performance, our goal is to assess how LLMs can contribute to diverse molecule tasks. We identify several classification and regression prediction tasks across six standard molecule datasets. Subsequently, we carefully design a set of prompts to query LLMs on these tasks and compare their performance with existing Machine Learning (ML) models, which include text-based models and those specifically designed for analysing the geometric structure of molecules. Our investigation reveals several key insights: Firstly, LLMs generally lag behind ML models in achieving competitive performance on molecule tasks, particularly when compared to models adept at capturing the geometric structure of molecules, highlighting the constrained ability of LLMs to comprehend graph data. Secondly, LLMs show promise in enhancing the performance of ML models when used collaboratively. Lastly, we engage in a discourse regarding the challenges and promising avenues to harness LLMs for molecule prediction tasks. The code and models are available at https://github.com/zhiqiangzhongddu/LLMaMol.
Harnessing Large Language Models as Post-hoc Correctors
Feb 20, 2024Abstract:As Machine Learning (ML) models grow in size and demand higher-quality training data, the expenses associated with re-training and fine-tuning these models are escalating rapidly. Inspired by recent impressive achievements of Large Language Models (LLMs) in different fields, this paper delves into the question: can LLMs efficiently improve an ML's performance at a minimal cost? We show that, through our proposed training-free framework LlmCorr, an LLM can work as a post-hoc corrector to propose corrections for the predictions of an arbitrary ML model. In particular, we form a contextual knowledge database by incorporating the dataset's label information and the ML model's predictions on the validation dataset. Leveraging the in-context learning capability of LLMs, we ask the LLM to summarise the instances in which the ML model makes mistakes and the correlation between primary predictions and true labels. Following this, the LLM can transfer its acquired knowledge to suggest corrections for the ML model's predictions. Our experimental results on the challenging molecular predictions show that LlmCorr improves the performance of a number of models by up to 39%.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge