David Chang
Local-Canonicalization Equivariant Graph Neural Networks for Sample-Efficient and Generalizable Swarm Robot Control
Sep 17, 2025Abstract:Multi-agent reinforcement learning (MARL) has emerged as a powerful paradigm for coordinating swarms of agents in complex decision-making, yet major challenges remain. In competitive settings such as pursuer-evader tasks, simultaneous adaptation can destabilize training; non-kinetic countermeasures often fail under adverse conditions; and policies trained in one configuration rarely generalize to environments with a different number of agents. To address these issues, we propose the Local-Canonicalization Equivariant Graph Neural Networks (LEGO) framework, which integrates seamlessly with popular MARL algorithms such as MAPPO. LEGO employs graph neural networks to capture permutation equivariance and generalization to different agent numbers, canonicalization to enforce E(n)-equivariance, and heterogeneous representations to encode role-specific inductive biases. Experiments on cooperative and competitive swarm benchmarks show that LEGO outperforms strong baselines and improves generalization. In real-world experiments, LEGO demonstrates robustness to varying team sizes and agent failure.
MedDistant19: A Challenging Benchmark for Distantly Supervised Biomedical Relation Extraction
Apr 10, 2022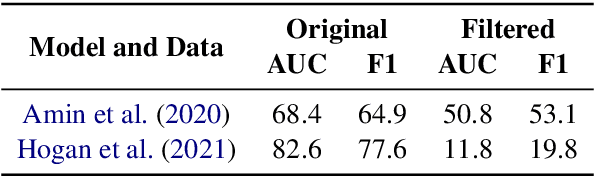
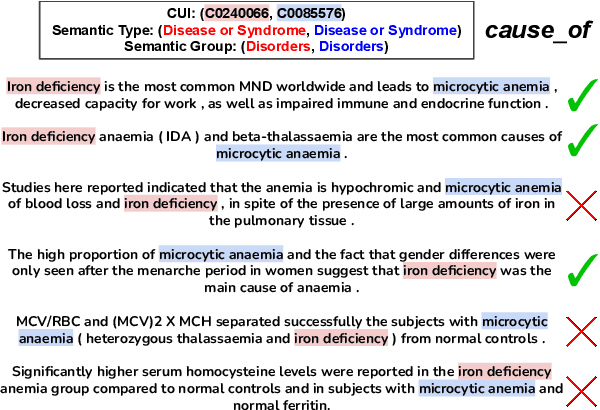

Abstract:Relation Extraction in the biomedical domain is challenging due to the lack of labeled data and high annotation costs, needing domain experts. Distant supervision is commonly used as a way to tackle the scarcity of annotated data by automatically pairing knowledge graph relationships with raw texts. Distantly Supervised Biomedical Relation Extraction (Bio-DSRE) models can seemingly produce very accurate results in several benchmarks. However, given the challenging nature of the task, we set out to investigate the validity of such impressive results. We probed the datasets used by Amin et al. (2020) and Hogan et al. (2021) and found a significant overlap between training and evaluation relationships that, once resolved, reduced the accuracy of the models by up to 71%. Furthermore, we noticed several inconsistencies with the data construction process, such as creating negative samples and improper handling of redundant relationships. We mitigate these issues and present MedDistant19, a new benchmark dataset obtained by aligning the MEDLINE abstracts with the widely used SNOMED Clinical Terms (SNOMED CT) knowledge base. We experimented with several state-of-the-art models achieving an AUC of 55.4% and 49.8% at sentence- and bag-level, showing that there is still plenty of room for improvement.
Neural Natural Language Processing for Unstructured Data in Electronic Health Records: a Review
Jul 07, 2021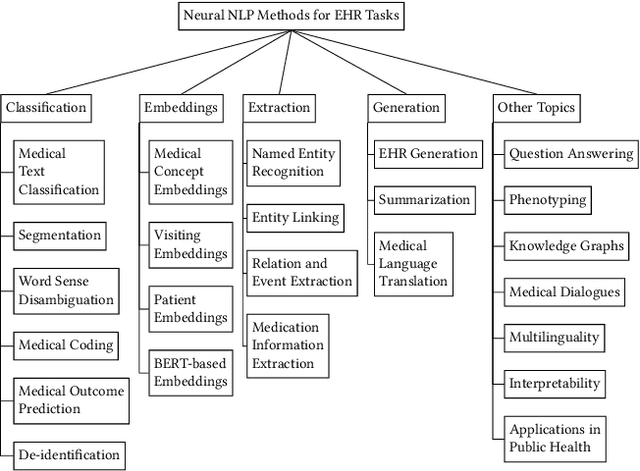
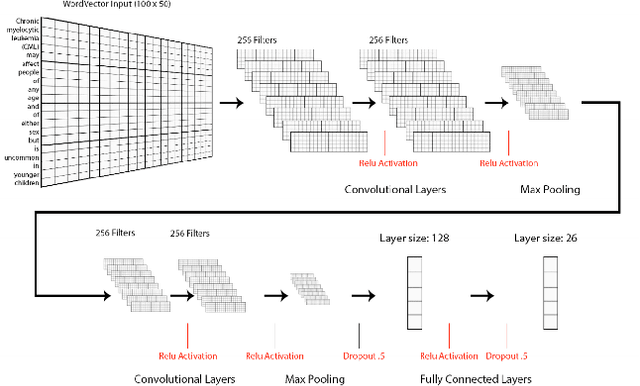
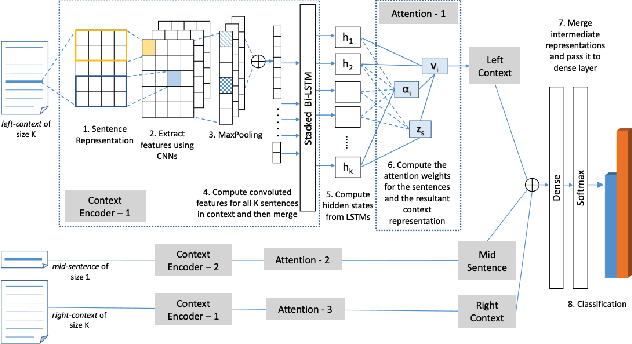

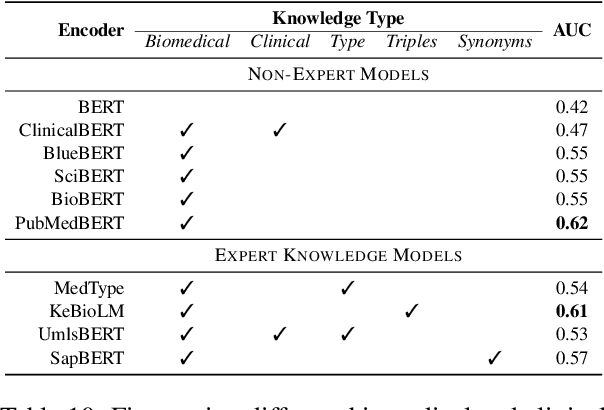
Abstract:Electronic health records (EHRs), digital collections of patient healthcare events and observations, are ubiquitous in medicine and critical to healthcare delivery, operations, and research. Despite this central role, EHRs are notoriously difficult to process automatically. Well over half of the information stored within EHRs is in the form of unstructured text (e.g. provider notes, operation reports) and remains largely untapped for secondary use. Recently, however, newer neural network and deep learning approaches to Natural Language Processing (NLP) have made considerable advances, outperforming traditional statistical and rule-based systems on a variety of tasks. In this survey paper, we summarize current neural NLP methods for EHR applications. We focus on a broad scope of tasks, namely, classification and prediction, word embeddings, extraction, generation, and other topics such as question answering, phenotyping, knowledge graphs, medical dialogue, multilinguality, interpretability, etc.
Benchmark and Best Practices for Biomedical Knowledge Graph Embeddings
Jun 24, 2020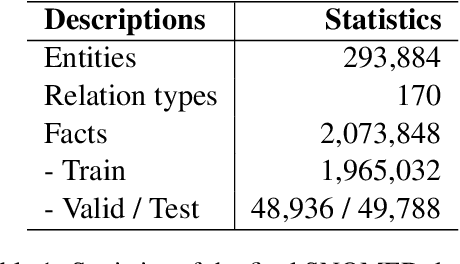
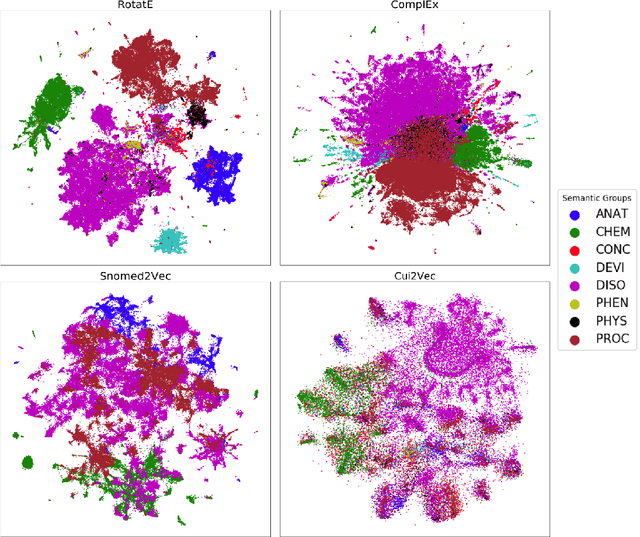
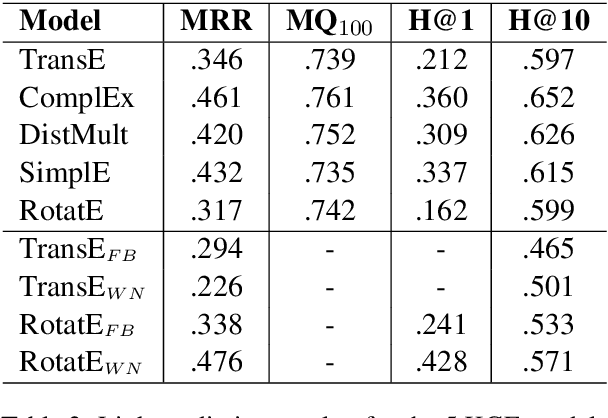
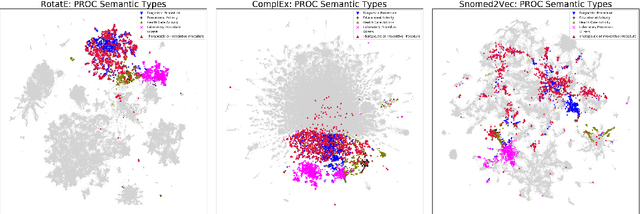
Abstract:Much of biomedical and healthcare data is encoded in discrete, symbolic form such as text and medical codes. There is a wealth of expert-curated biomedical domain knowledge stored in knowledge bases and ontologies, but the lack of reliable methods for learning knowledge representation has limited their usefulness in machine learning applications. While text-based representation learning has significantly improved in recent years through advances in natural language processing, attempts to learn biomedical concept embeddings so far have been lacking. A recent family of models called knowledge graph embeddings have shown promising results on general domain knowledge graphs, and we explore their capabilities in the biomedical domain. We train several state-of-the-art knowledge graph embedding models on the SNOMED-CT knowledge graph, provide a benchmark with comparison to existing methods and in-depth discussion on best practices, and make a case for the importance of leveraging the multi-relational nature of knowledge graphs for learning biomedical knowledge representation. The embeddings, code, and materials will be made available to the communitY.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge