Günter Neumann
Low-Resource Transliteration for Roman-Urdu and Urdu Using Transformer-Based Models
Mar 27, 2025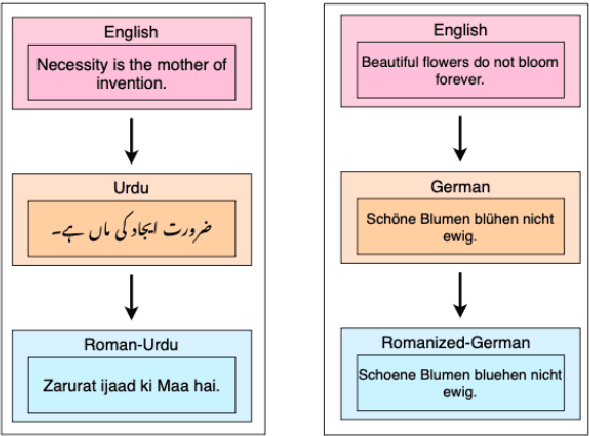
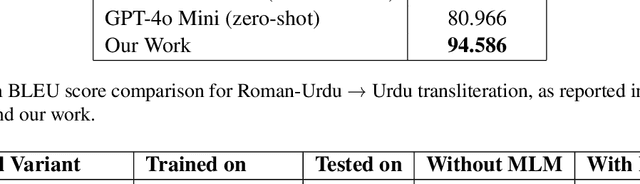
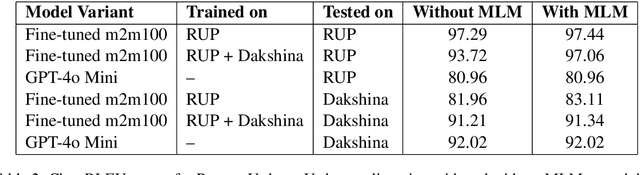
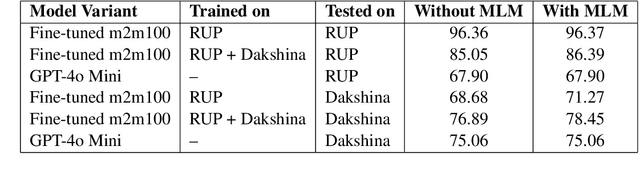
Abstract:As the Information Retrieval (IR) field increasingly recognizes the importance of inclusivity, addressing the needs of low-resource languages remains a significant challenge. Transliteration between Urdu and its Romanized form, Roman Urdu, remains underexplored despite the widespread use of both scripts in South Asia. Prior work using RNNs on the Roman-Urdu-Parl dataset showed promising results but suffered from poor domain adaptability and limited evaluation. We propose a transformer-based approach using the m2m100 multilingual translation model, enhanced with masked language modeling (MLM) pretraining and fine-tuning on both Roman-Urdu-Parl and the domain-diverse Dakshina dataset. To address previous evaluation flaws, we introduce rigorous dataset splits and assess performance using BLEU, character-level BLEU, and CHRF. Our model achieves strong transliteration performance, with Char-BLEU scores of 96.37 for Urdu->Roman-Urdu and 97.44 for Roman-Urdu->Urdu. These results outperform both RNN baselines and GPT-4o Mini and demonstrate the effectiveness of multilingual transfer learning for low-resource transliteration tasks.
Enabling Low-Resource Language Retrieval: Establishing Baselines for Urdu MS MARCO
Dec 17, 2024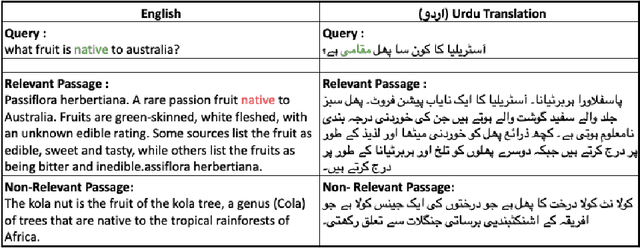
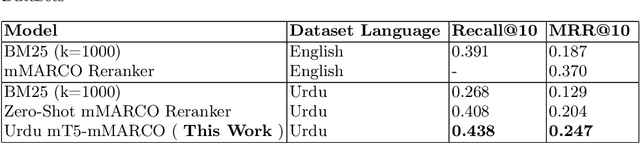
Abstract:As the Information Retrieval (IR) field increasingly recognizes the importance of inclusivity, addressing the needs of low-resource languages remains a significant challenge. This paper introduces the first large-scale Urdu IR dataset, created by translating the MS MARCO dataset through machine translation. We establish baseline results through zero-shot learning for IR in Urdu and subsequently apply the mMARCO multilingual IR methodology to this newly translated dataset. Our findings demonstrate that the fine-tuned model (Urdu-mT5-mMARCO) achieves a Mean Reciprocal Rank (MRR@10) of 0.247 and a Recall@10 of 0.439, representing significant improvements over zero-shot results and showing the potential for expanding IR access for Urdu speakers. By bridging access gaps for speakers of low-resource languages, this work not only advances multilingual IR research but also emphasizes the ethical and societal importance of inclusive IR technologies. This work provides valuable insights into the challenges and solutions for improving language representation and lays the groundwork for future research, especially in South Asian languages, which can benefit from the adaptable methods used in this study.
Temporal Knowledge Graph Reasoning with Low-rank and Model-agnostic Representations
Apr 10, 2022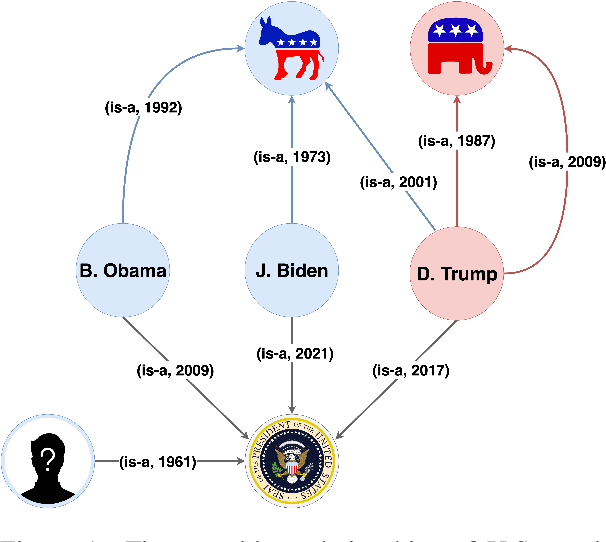
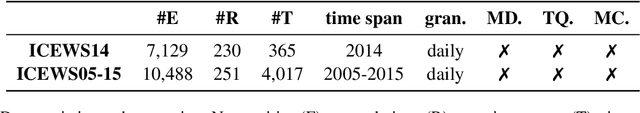
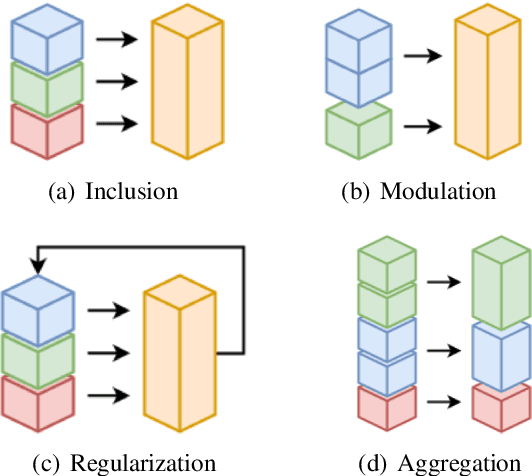
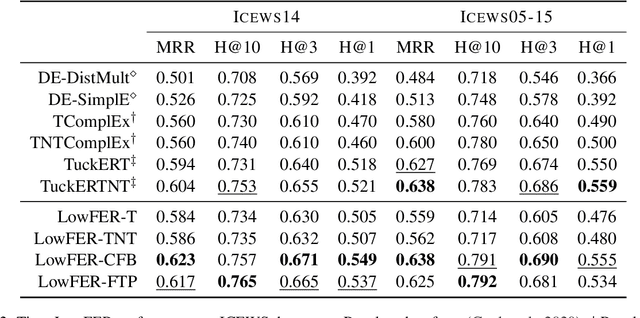
Abstract:Temporal knowledge graph completion (TKGC) has become a popular approach for reasoning over the event and temporal knowledge graphs, targeting the completion of knowledge with accurate but missing information. In this context, tensor decomposition has successfully modeled interactions between entities and relations. Their effectiveness in static knowledge graph completion motivates us to introduce Time-LowFER, a family of parameter-efficient and time-aware extensions of the low-rank tensor factorization model LowFER. Noting several limitations in current approaches to represent time, we propose a cycle-aware time-encoding scheme for time features, which is model-agnostic and offers a more generalized representation of time. We implement our methods in a unified temporal knowledge graph embedding framework, focusing on time-sensitive data processing. The experiments show that our proposed methods perform on par or better than the state-of-the-art semantic matching models on two benchmarks.
MedDistant19: A Challenging Benchmark for Distantly Supervised Biomedical Relation Extraction
Apr 10, 2022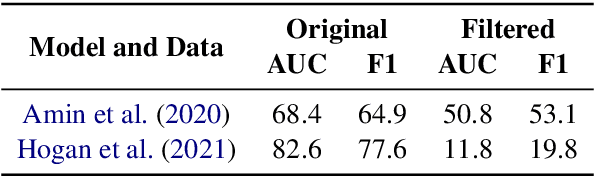
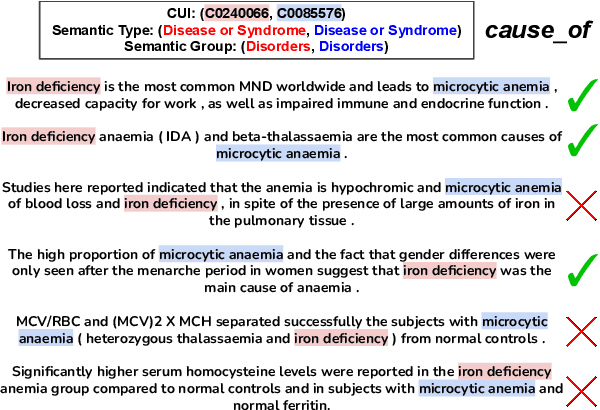
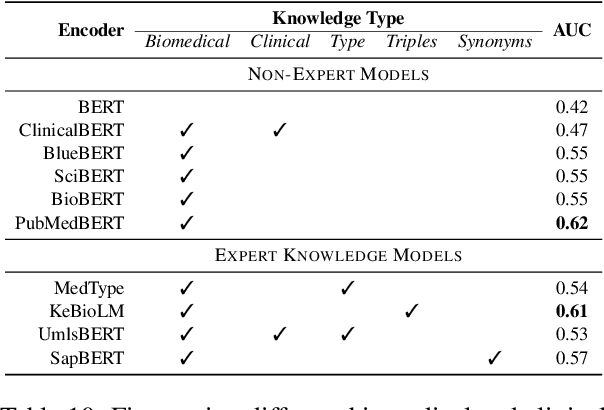

Abstract:Relation Extraction in the biomedical domain is challenging due to the lack of labeled data and high annotation costs, needing domain experts. Distant supervision is commonly used as a way to tackle the scarcity of annotated data by automatically pairing knowledge graph relationships with raw texts. Distantly Supervised Biomedical Relation Extraction (Bio-DSRE) models can seemingly produce very accurate results in several benchmarks. However, given the challenging nature of the task, we set out to investigate the validity of such impressive results. We probed the datasets used by Amin et al. (2020) and Hogan et al. (2021) and found a significant overlap between training and evaluation relationships that, once resolved, reduced the accuracy of the models by up to 71%. Furthermore, we noticed several inconsistencies with the data construction process, such as creating negative samples and improper handling of redundant relationships. We mitigate these issues and present MedDistant19, a new benchmark dataset obtained by aligning the MEDLINE abstracts with the widely used SNOMED Clinical Terms (SNOMED CT) knowledge base. We experimented with several state-of-the-art models achieving an AUC of 55.4% and 49.8% at sentence- and bag-level, showing that there is still plenty of room for improvement.
Few-Shot Cross-lingual Transfer for Coarse-grained De-identification of Code-Mixed Clinical Texts
Apr 10, 2022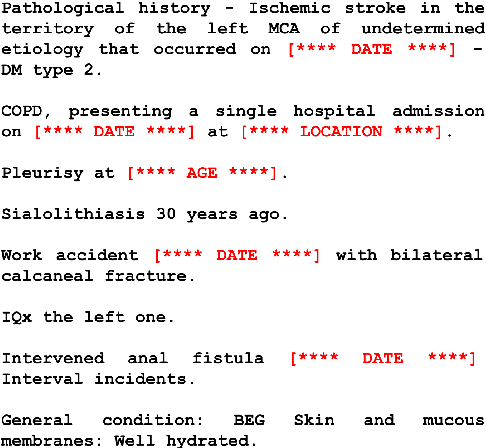
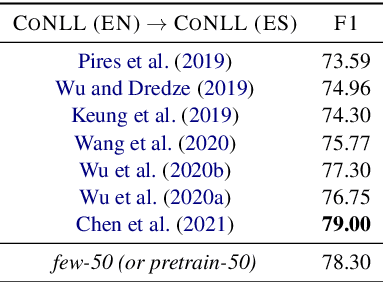
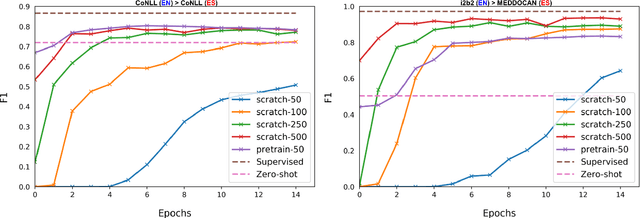
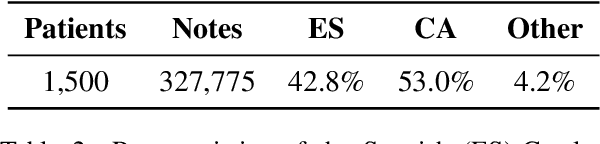
Abstract:Despite the advances in digital healthcare systems offering curated structured knowledge, much of the critical information still lies in large volumes of unlabeled and unstructured clinical texts. These texts, which often contain protected health information (PHI), are exposed to information extraction tools for downstream applications, risking patient identification. Existing works in de-identification rely on using large-scale annotated corpora in English, which often are not suitable in real-world multilingual settings. Pre-trained language models (LM) have shown great potential for cross-lingual transfer in low-resource settings. In this work, we empirically show the few-shot cross-lingual transfer property of LMs for named entity recognition (NER) and apply it to solve a low-resource and real-world challenge of code-mixed (Spanish-Catalan) clinical notes de-identification in the stroke domain. We annotate a gold evaluation dataset to assess few-shot setting performance where we only use a few hundred labeled examples for training. Our model improves the zero-shot F1-score from 73.7% to 91.2% on the gold evaluation set when adapting Multilingual BERT (mBERT) (Devlin et al., 2019) from the MEDDOCAN (Marimon et al., 2019) corpus with our few-shot cross-lingual target corpus. When generalized to an out-of-sample test set, the best model achieves a human-evaluation F1-score of 97.2%.
LowFER: Low-rank Bilinear Pooling for Link Prediction
Aug 25, 2020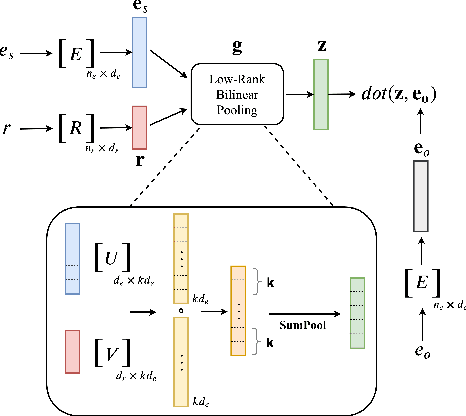
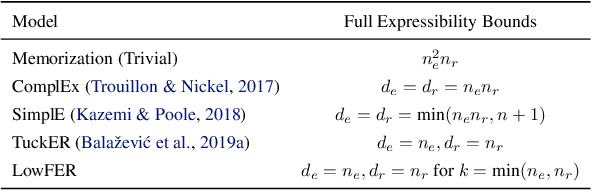
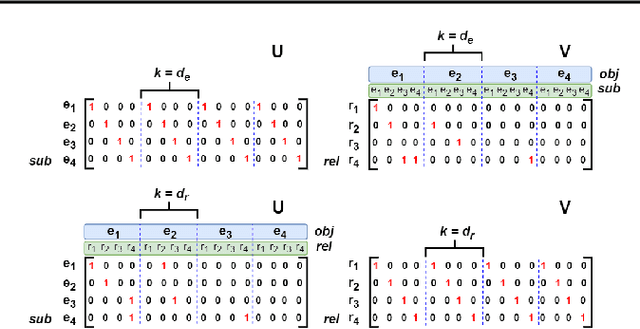

Abstract:Knowledge graphs are incomplete by nature, with only a limited number of observed facts from the world knowledge being represented as structured relations between entities. To partly address this issue, an important task in statistical relational learning is that of link prediction or knowledge graph completion. Both linear and non-linear models have been proposed to solve the problem. Bilinear models, while expressive, are prone to overfitting and lead to quadratic growth of parameters in number of relations. Simpler models have become more standard, with certain constraints on bilinear map as relation parameters. In this work, we propose a factorized bilinear pooling model, commonly used in multi-modal learning, for better fusion of entities and relations, leading to an efficient and constraint-free model. We prove that our model is fully expressive, providing bounds on the embedding dimensionality and factorization rank. Our model naturally generalizes Tucker decomposition based TuckER model, which has been shown to generalize other models, as efficient low-rank approximation without substantially compromising the performance. Due to low-rank approximation, the model complexity can be controlled by the factorization rank, avoiding the possible cubic growth of TuckER. Empirically, we evaluate on real-world datasets, reaching on par or state-of-the-art performance. At extreme low-ranks, model preserves the performance while staying parameter efficient.
A Data-driven Approach for Noise Reduction in Distantly Supervised Biomedical Relation Extraction
May 26, 2020

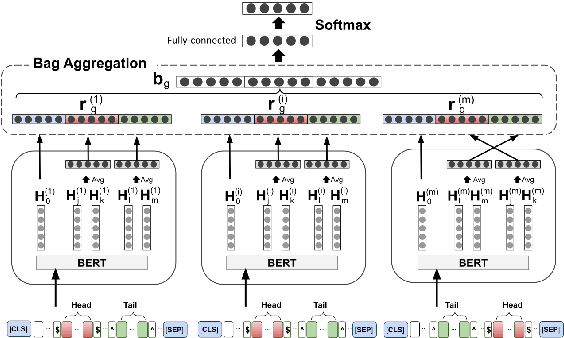

Abstract:Fact triples are a common form of structured knowledge used within the biomedical domain. As the amount of unstructured scientific texts continues to grow, manual annotation of these texts for the task of relation extraction becomes increasingly expensive. Distant supervision offers a viable approach to combat this by quickly producing large amounts of labeled, but considerably noisy, data. We aim to reduce such noise by extending an entity-enriched relation classification BERT model to the problem of multiple instance learning, and defining a simple data encoding scheme that significantly reduces noise, reaching state-of-the-art performance for distantly-supervised biomedical relation extraction. Our approach further encodes knowledge about the direction of relation triples, allowing for increased focus on relation learning by reducing noise and alleviating the need for joint learning with knowledge graph completion.
LightRel SemEval-2018 Task 7: Lightweight and Fast Relation Classification
Apr 19, 2018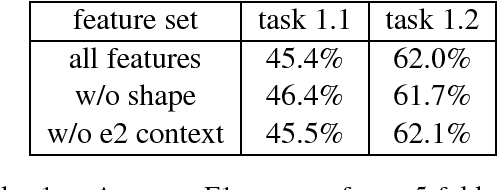
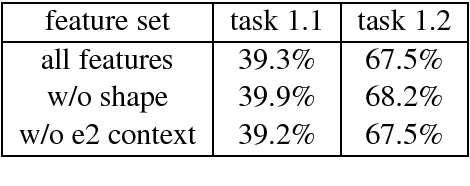
Abstract:We present LightRel, a lightweight and fast relation classifier. Our goal is to develop a high baseline for different relation extraction tasks. By defining only very few data-internal, word-level features and external knowledge sources in the form of word clusters and word embeddings, we train a fast and simple linear classifier.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge