Katherine Ann Dunfield
LowFER: Low-rank Bilinear Pooling for Link Prediction
Aug 25, 2020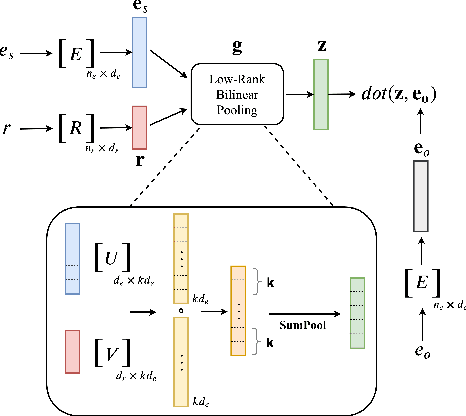
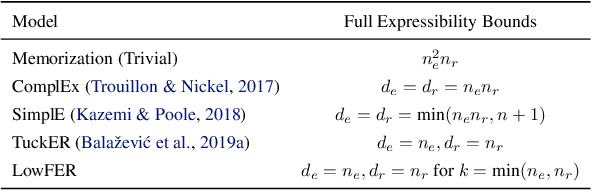
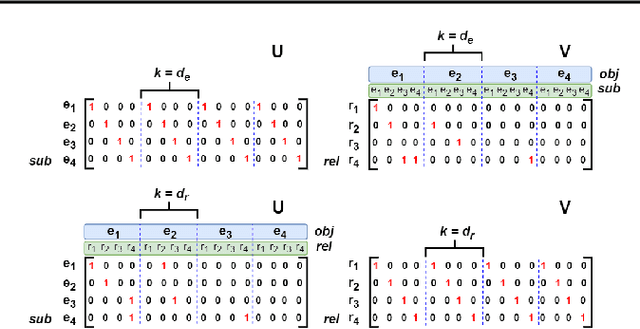

Abstract:Knowledge graphs are incomplete by nature, with only a limited number of observed facts from the world knowledge being represented as structured relations between entities. To partly address this issue, an important task in statistical relational learning is that of link prediction or knowledge graph completion. Both linear and non-linear models have been proposed to solve the problem. Bilinear models, while expressive, are prone to overfitting and lead to quadratic growth of parameters in number of relations. Simpler models have become more standard, with certain constraints on bilinear map as relation parameters. In this work, we propose a factorized bilinear pooling model, commonly used in multi-modal learning, for better fusion of entities and relations, leading to an efficient and constraint-free model. We prove that our model is fully expressive, providing bounds on the embedding dimensionality and factorization rank. Our model naturally generalizes Tucker decomposition based TuckER model, which has been shown to generalize other models, as efficient low-rank approximation without substantially compromising the performance. Due to low-rank approximation, the model complexity can be controlled by the factorization rank, avoiding the possible cubic growth of TuckER. Empirically, we evaluate on real-world datasets, reaching on par or state-of-the-art performance. At extreme low-ranks, model preserves the performance while staying parameter efficient.
A Data-driven Approach for Noise Reduction in Distantly Supervised Biomedical Relation Extraction
May 26, 2020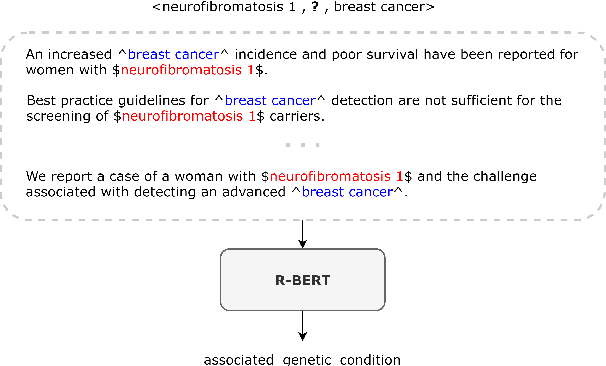

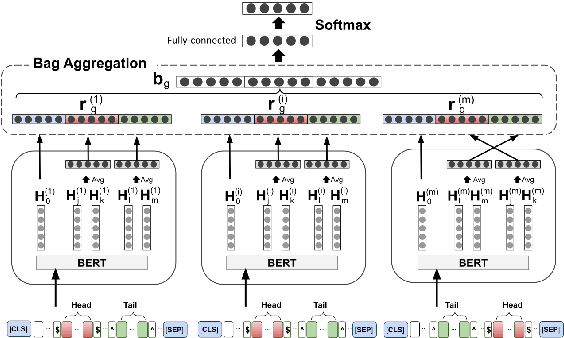

Abstract:Fact triples are a common form of structured knowledge used within the biomedical domain. As the amount of unstructured scientific texts continues to grow, manual annotation of these texts for the task of relation extraction becomes increasingly expensive. Distant supervision offers a viable approach to combat this by quickly producing large amounts of labeled, but considerably noisy, data. We aim to reduce such noise by extending an entity-enriched relation classification BERT model to the problem of multiple instance learning, and defining a simple data encoding scheme that significantly reduces noise, reaching state-of-the-art performance for distantly-supervised biomedical relation extraction. Our approach further encodes knowledge about the direction of relation triples, allowing for increased focus on relation learning by reducing noise and alleviating the need for joint learning with knowledge graph completion.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge