Darren Dancey
A Novel Spike Transformer Network for Depth Estimation from Event Cameras via Cross-modality Knowledge Distillation
May 01, 2024Abstract:Depth estimation is crucial for interpreting complex environments, especially in areas such as autonomous vehicle navigation and robotics. Nonetheless, obtaining accurate depth readings from event camera data remains a formidable challenge. Event cameras operate differently from traditional digital cameras, continuously capturing data and generating asynchronous binary spikes that encode time, location, and light intensity. Yet, the unique sampling mechanisms of event cameras render standard image based algorithms inadequate for processing spike data. This necessitates the development of innovative, spike-aware algorithms tailored for event cameras, a task compounded by the irregularity, continuity, noise, and spatial and temporal characteristics inherent in spiking data.Harnessing the strong generalization capabilities of transformer neural networks for spatiotemporal data, we propose a purely spike-driven spike transformer network for depth estimation from spiking camera data. To address performance limitations with Spiking Neural Networks (SNN), we introduce a novel single-stage cross-modality knowledge transfer framework leveraging knowledge from a large vision foundational model of artificial neural networks (ANN) (DINOv2) to enhance the performance of SNNs with limited data. Our experimental results on both synthetic and real datasets show substantial improvements over existing models, with notable gains in Absolute Relative and Square Relative errors (49% and 39.77% improvements over the benchmark model Spike-T, respectively). Besides accuracy, the proposed model also demonstrates reduced power consumptions, a critical factor for practical applications.
A Fast Fourier Convolutional Deep Neural Network For Accurate and Explainable Discrimination Of Wheat Yellow Rust And Nitrogen Deficiency From Sentinel-2 Time-Series Data
Jun 29, 2023Abstract:Accurate and timely detection of plant stress is essential for yield protection, allowing better-targeted intervention strategies. Recent advances in remote sensing and deep learning have shown great potential for rapid non-invasive detection of plant stress in a fully automated and reproducible manner. However, the existing models always face several challenges: 1) computational inefficiency and the misclassifications between the different stresses with similar symptoms; and 2) the poor interpretability of the host-stress interaction. In this work, we propose a novel fast Fourier Convolutional Neural Network (FFDNN) for accurate and explainable detection of two plant stresses with similar symptoms (i.e. Wheat Yellow Rust And Nitrogen Deficiency). Specifically, unlike the existing CNN models, the main components of the proposed model include: 1) a fast Fourier convolutional block, a newly fast Fourier transformation kernel as the basic perception unit, to substitute the traditional convolutional kernel to capture both local and global responses to plant stress in various time-scale and improve computing efficiency with reduced learning parameters in Fourier domain; 2) Capsule Feature Encoder to encapsulate the extracted features into a series of vector features to represent part-to-whole relationship with the hierarchical structure of the host-stress interactions of the specific stress. In addition, in order to alleviate over-fitting, a photochemical vegetation indices-based filter is placed as pre-processing operator to remove the non-photochemical noises from the input Sentinel-2 time series.
sMRI-PatchNet: A novel explainable patch-based deep learning network for Alzheimer's disease diagnosis and discriminative atrophy localisation with Structural MRI
Feb 20, 2023Abstract:Structural magnetic resonance imaging (sMRI) can identify subtle brain changes due to its high contrast for soft tissues and high spatial resolution. It has been widely used in diagnosing neurological brain diseases, such as Alzheimer disease (AD). However, the size of 3D high-resolution data poses a significant challenge for data analysis and processing. Since only a few areas of the brain show structural changes highly associated with AD, the patch-based methods dividing the whole image data into several small regular patches have shown promising for more efficient sMRI-based image analysis. The major challenges of the patch-based methods on sMRI include identifying the discriminative patches, combining features from the discrete discriminative patches, and designing appropriate classifiers. This work proposes a novel patch-based deep learning network (sMRI-PatchNet) with explainable patch localisation and selection for AD diagnosis using sMRI. Specifically, it consists of two primary components: 1) A fast and efficient explainable patch selection mechanism for determining the most discriminative patches based on computing the SHapley Additive exPlanations (SHAP) contribution to a transfer learning model for AD diagnosis on massive medical data; and 2) A novel patch-based network for extracting deep features and AD classfication from the selected patches with position embeddings to retain position information, capable of capturing the global and local information of inter- and intra-patches. This method has been applied for the AD classification and the prediction of the transitional state moderate cognitive impairment (MCI) conversion with real datasets.
Layer-Wise Partitioning and Merging for Efficient and Scalable Deep Learning
Jul 22, 2022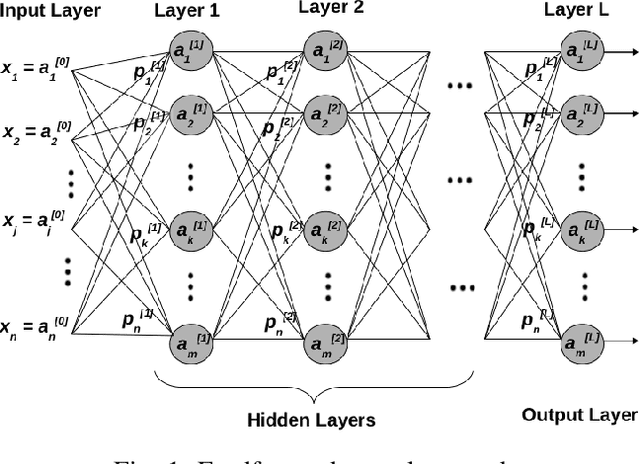
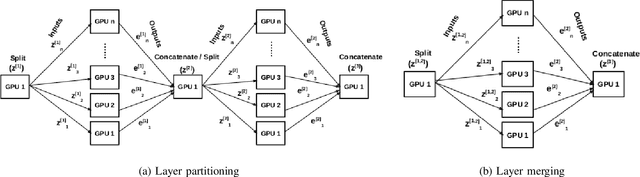
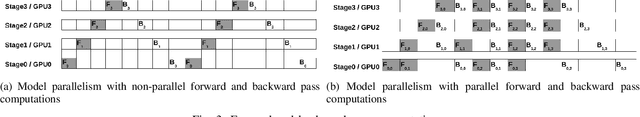
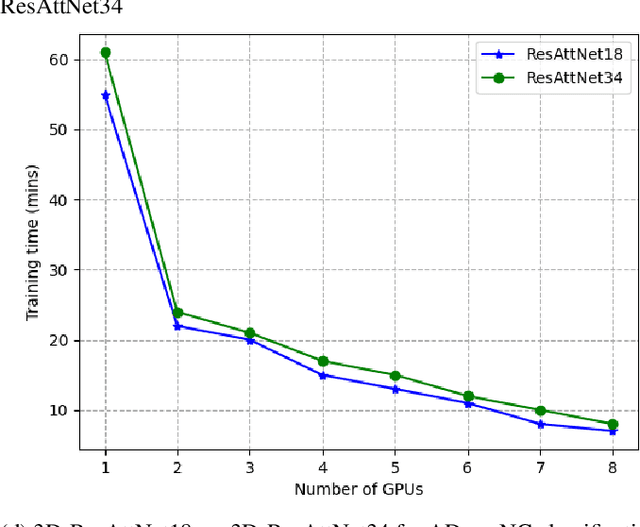
Abstract:Deep Neural Network (DNN) models are usually trained sequentially from one layer to another, which causes forward, backward and update locking's problems, leading to poor performance in terms of training time. The existing parallel strategies to mitigate these problems provide suboptimal runtime performance. In this work, we have proposed a novel layer-wise partitioning and merging, forward and backward pass parallel framework to provide better training performance. The novelty of the proposed work consists of 1) a layer-wise partition and merging model which can minimise communication overhead between devices without the memory cost of existing strategies during the training process; 2) a forward pass and backward pass parallelisation and optimisation to address the update locking problem and minimise the total training cost. The experimental evaluation on real use cases shows that the proposed method outperforms the state-of-the-art approaches in terms of training speed; and achieves almost linear speedup without compromising the accuracy performance of the non-parallel approach.
A Latent Encoder Coupled Generative Adversarial Network (LE-GAN) for Efficient Hyperspectral Image Super-resolution
Nov 16, 2021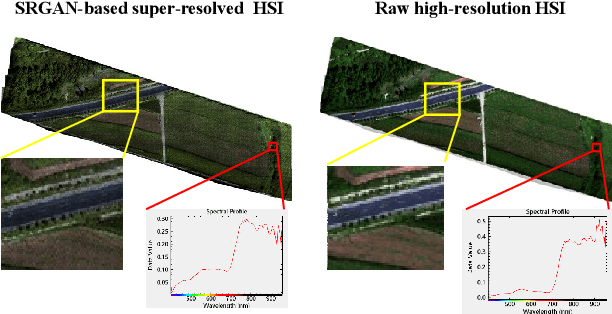
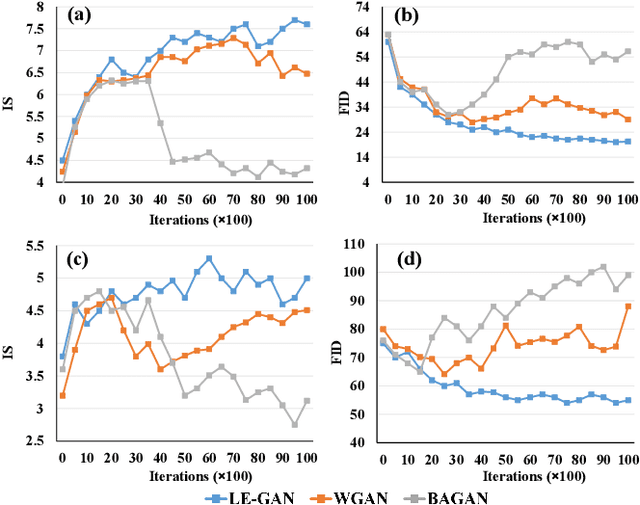
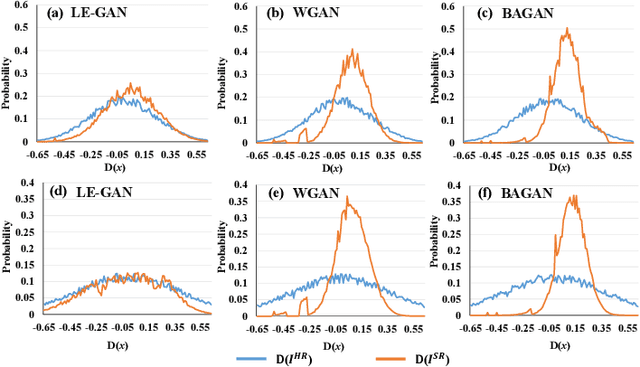
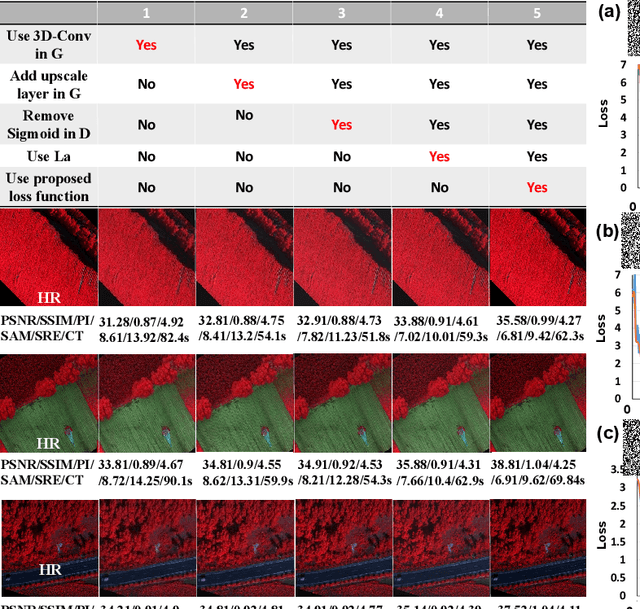
Abstract:Realistic hyperspectral image (HSI) super-resolution (SR) techniques aim to generate a high-resolution (HR) HSI with higher spectral and spatial fidelity from its low-resolution (LR) counterpart. The generative adversarial network (GAN) has proven to be an effective deep learning framework for image super-resolution. However, the optimisation process of existing GAN-based models frequently suffers from the problem of mode collapse, leading to the limited capacity of spectral-spatial invariant reconstruction. This may cause the spectral-spatial distortion on the generated HSI, especially with a large upscaling factor. To alleviate the problem of mode collapse, this work has proposed a novel GAN model coupled with a latent encoder (LE-GAN), which can map the generated spectral-spatial features from the image space to the latent space and produce a coupling component to regularise the generated samples. Essentially, we treat an HSI as a high-dimensional manifold embedded in a latent space. Thus, the optimisation of GAN models is converted to the problem of learning the distributions of high-resolution HSI samples in the latent space, making the distributions of the generated super-resolution HSIs closer to those of their original high-resolution counterparts. We have conducted experimental evaluations on the model performance of super-resolution and its capability in alleviating mode collapse. The proposed approach has been tested and validated based on two real HSI datasets with different sensors (i.e. AVIRIS and UHD-185) for various upscaling factors and added noise levels, and compared with the state-of-the-art super-resolution models (i.e. HyCoNet, LTTR, BAGAN, SR- GAN, WGAN).
A Biologically Interpretable Two-stage Deep Neural Network (BIT-DNN) For Hyperspectral Imagery Classification
Apr 19, 2020

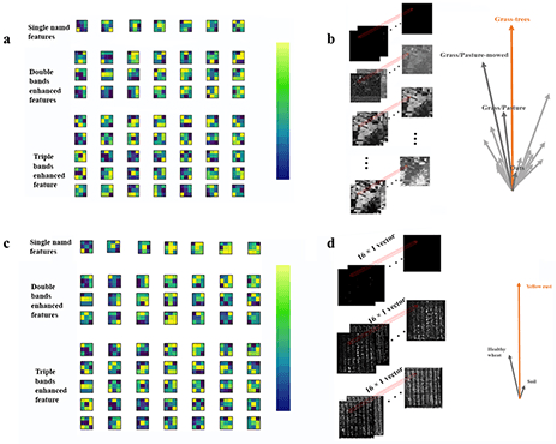
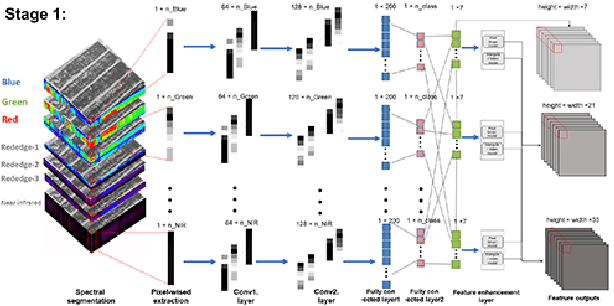
Abstract:Spectral-spatial based deep learning models have recently proven to be effective in hyperspectral image (HSI) classification for various earth monitoring applications such as land cover classification and agricultural monitoring. However, due to the nature of "black-box" model representation, how to explain and interpret the learning process and the model decision remains an open problem. This study proposes an interpretable deep learning model -- a biologically interpretable two-stage deep neural network (BIT-DNN), by integrating biochemical and biophysical associated information into the proposed framework, capable of achieving both high accuracy and interpretability on HSI based classification tasks. The proposed model introduces a two-stage feature learning process. In the first stage, an enhanced interpretable feature block extracts low-level spectral features associated with the biophysical and biochemical attributes of the target entities; and in the second stage, an interpretable capsule block extracts and encapsulates the high-level joint spectral-spatial features into the featured tensors representing the hierarchical structure of the biophysical and biochemical attributes of the target ground entities, which provides the model an improved performance on classification and intrinsic interpretability. We have tested and evaluated the model using two real HSI datasets for crop type recognition and crop disease recognition tasks and compared it with six state-of-the-art machine learning models. The results demonstrate that the proposed model has competitive advantages in terms of both classification accuracy and model interpretability.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge