Lianghao Han
WaveDiffUR: A diffusion SDE-based solver for ultra magnification super-resolution in remote sensing images
Dec 25, 2024Abstract:Deep neural networks have recently achieved significant advancements in remote sensing superresolu-tion (SR). However, most existing methods are limited to low magnification rates (e.g., 2 or 4) due to the escalating ill-posedness at higher magnification scales. To tackle this challenge, we redefine high-magnification SR as the ultra-resolution (UR) problem, reframing it as solving a conditional diffusion stochastic differential equation (SDE). In this context, we propose WaveDiffUR, a novel wavelet-domain diffusion UR solver that decomposes the UR process into sequential sub-processes addressing conditional wavelet components. WaveDiffUR iteratively reconstructs low-frequency wavelet details (ensuring global consistency) and high-frequency components (enhancing local fidelity) by incorporating pre-trained SR models as plug-and-play modules. This modularity mitigates the ill-posedness of the SDE and ensures scalability across diverse applications. To address limitations in fixed boundary conditions at extreme magnifications, we introduce the cross-scale pyramid (CSP) constraint, a dynamic and adaptive framework that guides WaveDiffUR in generating fine-grained wavelet details, ensuring consistent and high-fidelity outputs even at extreme magnification rates.
A Novel Spike Transformer Network for Depth Estimation from Event Cameras via Cross-modality Knowledge Distillation
May 01, 2024Abstract:Depth estimation is crucial for interpreting complex environments, especially in areas such as autonomous vehicle navigation and robotics. Nonetheless, obtaining accurate depth readings from event camera data remains a formidable challenge. Event cameras operate differently from traditional digital cameras, continuously capturing data and generating asynchronous binary spikes that encode time, location, and light intensity. Yet, the unique sampling mechanisms of event cameras render standard image based algorithms inadequate for processing spike data. This necessitates the development of innovative, spike-aware algorithms tailored for event cameras, a task compounded by the irregularity, continuity, noise, and spatial and temporal characteristics inherent in spiking data.Harnessing the strong generalization capabilities of transformer neural networks for spatiotemporal data, we propose a purely spike-driven spike transformer network for depth estimation from spiking camera data. To address performance limitations with Spiking Neural Networks (SNN), we introduce a novel single-stage cross-modality knowledge transfer framework leveraging knowledge from a large vision foundational model of artificial neural networks (ANN) (DINOv2) to enhance the performance of SNNs with limited data. Our experimental results on both synthetic and real datasets show substantial improvements over existing models, with notable gains in Absolute Relative and Square Relative errors (49% and 39.77% improvements over the benchmark model Spike-T, respectively). Besides accuracy, the proposed model also demonstrates reduced power consumptions, a critical factor for practical applications.
sMRI-PatchNet: A novel explainable patch-based deep learning network for Alzheimer's disease diagnosis and discriminative atrophy localisation with Structural MRI
Feb 20, 2023Abstract:Structural magnetic resonance imaging (sMRI) can identify subtle brain changes due to its high contrast for soft tissues and high spatial resolution. It has been widely used in diagnosing neurological brain diseases, such as Alzheimer disease (AD). However, the size of 3D high-resolution data poses a significant challenge for data analysis and processing. Since only a few areas of the brain show structural changes highly associated with AD, the patch-based methods dividing the whole image data into several small regular patches have shown promising for more efficient sMRI-based image analysis. The major challenges of the patch-based methods on sMRI include identifying the discriminative patches, combining features from the discrete discriminative patches, and designing appropriate classifiers. This work proposes a novel patch-based deep learning network (sMRI-PatchNet) with explainable patch localisation and selection for AD diagnosis using sMRI. Specifically, it consists of two primary components: 1) A fast and efficient explainable patch selection mechanism for determining the most discriminative patches based on computing the SHapley Additive exPlanations (SHAP) contribution to a transfer learning model for AD diagnosis on massive medical data; and 2) A novel patch-based network for extracting deep features and AD classfication from the selected patches with position embeddings to retain position information, capable of capturing the global and local information of inter- and intra-patches. This method has been applied for the AD classification and the prediction of the transitional state moderate cognitive impairment (MCI) conversion with real datasets.
A Latent Encoder Coupled Generative Adversarial Network (LE-GAN) for Efficient Hyperspectral Image Super-resolution
Nov 16, 2021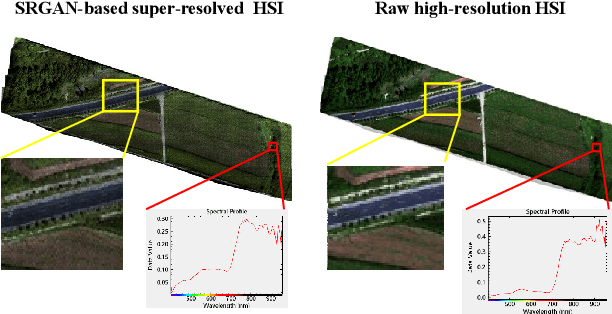
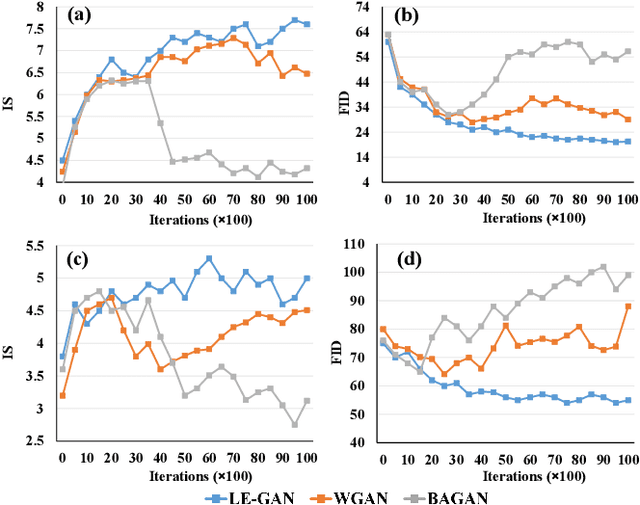
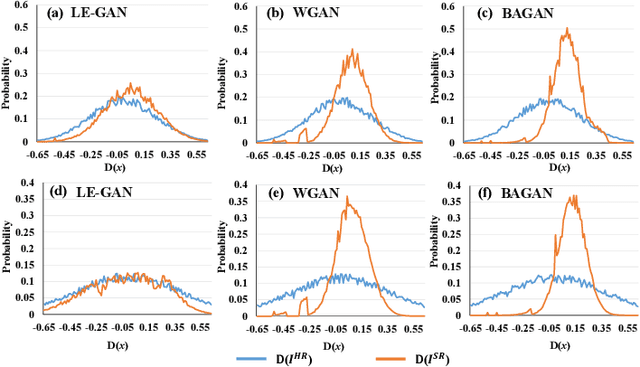
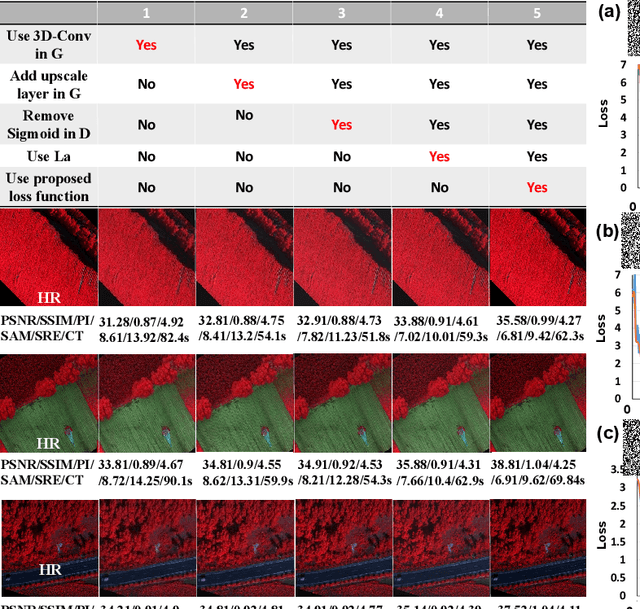
Abstract:Realistic hyperspectral image (HSI) super-resolution (SR) techniques aim to generate a high-resolution (HR) HSI with higher spectral and spatial fidelity from its low-resolution (LR) counterpart. The generative adversarial network (GAN) has proven to be an effective deep learning framework for image super-resolution. However, the optimisation process of existing GAN-based models frequently suffers from the problem of mode collapse, leading to the limited capacity of spectral-spatial invariant reconstruction. This may cause the spectral-spatial distortion on the generated HSI, especially with a large upscaling factor. To alleviate the problem of mode collapse, this work has proposed a novel GAN model coupled with a latent encoder (LE-GAN), which can map the generated spectral-spatial features from the image space to the latent space and produce a coupling component to regularise the generated samples. Essentially, we treat an HSI as a high-dimensional manifold embedded in a latent space. Thus, the optimisation of GAN models is converted to the problem of learning the distributions of high-resolution HSI samples in the latent space, making the distributions of the generated super-resolution HSIs closer to those of their original high-resolution counterparts. We have conducted experimental evaluations on the model performance of super-resolution and its capability in alleviating mode collapse. The proposed approach has been tested and validated based on two real HSI datasets with different sensors (i.e. AVIRIS and UHD-185) for various upscaling factors and added noise levels, and compared with the state-of-the-art super-resolution models (i.e. HyCoNet, LTTR, BAGAN, SR- GAN, WGAN).
CXR-Net: An Encoder-Decoder-Encoder Multitask Deep Neural Network for Explainable and Accurate Diagnosis of COVID-19 pneumonia with Chest X-ray Images
Oct 20, 2021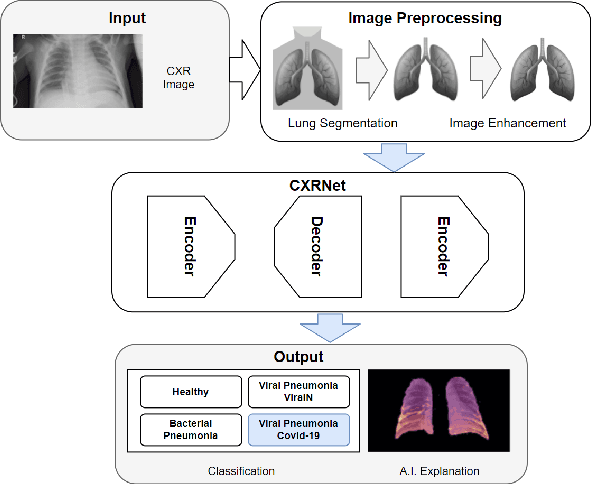
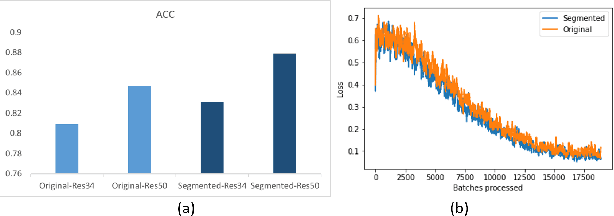
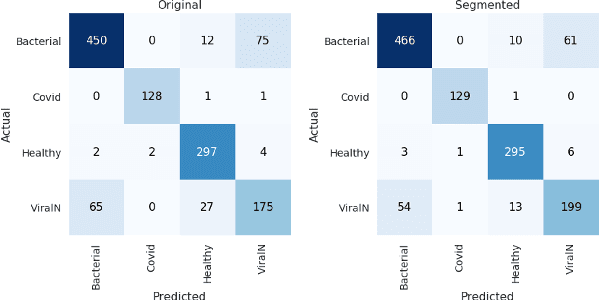
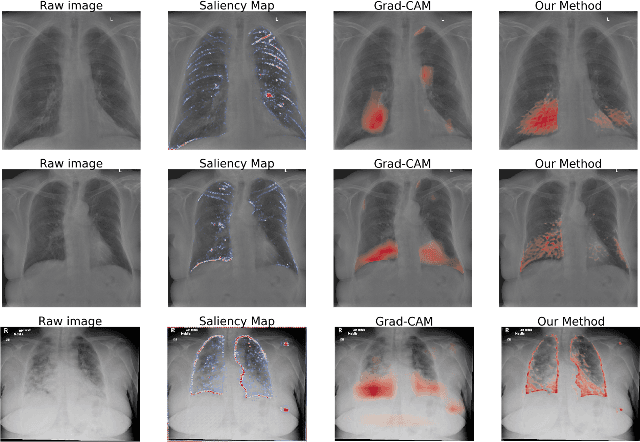
Abstract:Accurate and rapid detection of COVID-19 pneumonia is crucial for optimal patient treatment. Chest X-Ray (CXR) is the first line imaging test for COVID-19 pneumonia diagnosis as it is fast, cheap and easily accessible. Inspired by the success of deep learning (DL) in computer vision, many DL-models have been proposed to detect COVID-19 pneumonia using CXR images. Unfortunately, these deep classifiers lack the transparency in interpreting findings, which may limit their applications in clinical practice. The existing commonly used visual explanation methods are either too noisy or imprecise, with low resolution, and hence are unsuitable for diagnostic purposes. In this work, we propose a novel explainable deep learning framework (CXRNet) for accurate COVID-19 pneumonia detection with an enhanced pixel-level visual explanation from CXR images. The proposed framework is based on a new Encoder-Decoder-Encoder multitask architecture, allowing for both disease classification and visual explanation. The method has been evaluated on real world CXR datasets from both public and private data sources, including: healthy, bacterial pneumonia, viral pneumonia and COVID-19 pneumonia cases The experimental results demonstrate that the proposed method can achieve a satisfactory level of accuracy and provide fine-resolution classification activation maps for visual explanation in lung disease detection. The Average Accuracy, the Precision, Recall and F1-score of COVID-19 pneumonia reached 0.879, 0.985, 0.992 and 0.989, respectively. We have also found that using lung segmented (CXR) images can help improve the performance of the model. The proposed method can provide more detailed high resolution visual explanation for the classification decision, compared to current state-of-the-art visual explanation methods and has a great potential to be used in clinical practice for COVID-19 pneumonia diagnosis.
A Biologically Interpretable Two-stage Deep Neural Network (BIT-DNN) For Hyperspectral Imagery Classification
Apr 19, 2020

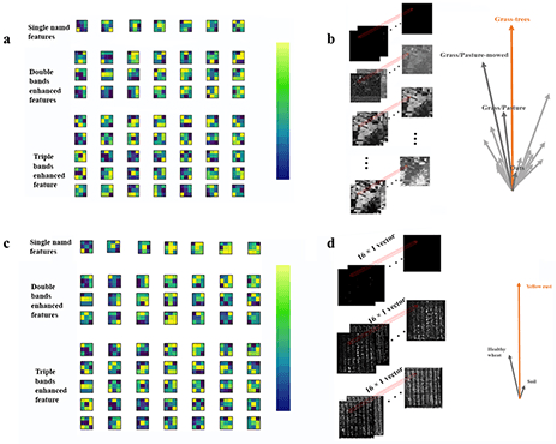
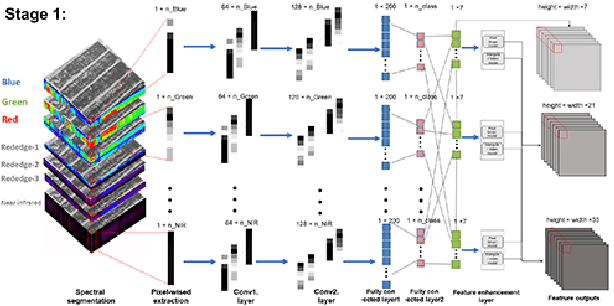
Abstract:Spectral-spatial based deep learning models have recently proven to be effective in hyperspectral image (HSI) classification for various earth monitoring applications such as land cover classification and agricultural monitoring. However, due to the nature of "black-box" model representation, how to explain and interpret the learning process and the model decision remains an open problem. This study proposes an interpretable deep learning model -- a biologically interpretable two-stage deep neural network (BIT-DNN), by integrating biochemical and biophysical associated information into the proposed framework, capable of achieving both high accuracy and interpretability on HSI based classification tasks. The proposed model introduces a two-stage feature learning process. In the first stage, an enhanced interpretable feature block extracts low-level spectral features associated with the biophysical and biochemical attributes of the target entities; and in the second stage, an interpretable capsule block extracts and encapsulates the high-level joint spectral-spatial features into the featured tensors representing the hierarchical structure of the biophysical and biochemical attributes of the target ground entities, which provides the model an improved performance on classification and intrinsic interpretability. We have tested and evaluated the model using two real HSI datasets for crop type recognition and crop disease recognition tasks and compared it with six state-of-the-art machine learning models. The results demonstrate that the proposed model has competitive advantages in terms of both classification accuracy and model interpretability.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge