Wenjiang Huang
A Fast Fourier Convolutional Deep Neural Network For Accurate and Explainable Discrimination Of Wheat Yellow Rust And Nitrogen Deficiency From Sentinel-2 Time-Series Data
Jun 29, 2023Abstract:Accurate and timely detection of plant stress is essential for yield protection, allowing better-targeted intervention strategies. Recent advances in remote sensing and deep learning have shown great potential for rapid non-invasive detection of plant stress in a fully automated and reproducible manner. However, the existing models always face several challenges: 1) computational inefficiency and the misclassifications between the different stresses with similar symptoms; and 2) the poor interpretability of the host-stress interaction. In this work, we propose a novel fast Fourier Convolutional Neural Network (FFDNN) for accurate and explainable detection of two plant stresses with similar symptoms (i.e. Wheat Yellow Rust And Nitrogen Deficiency). Specifically, unlike the existing CNN models, the main components of the proposed model include: 1) a fast Fourier convolutional block, a newly fast Fourier transformation kernel as the basic perception unit, to substitute the traditional convolutional kernel to capture both local and global responses to plant stress in various time-scale and improve computing efficiency with reduced learning parameters in Fourier domain; 2) Capsule Feature Encoder to encapsulate the extracted features into a series of vector features to represent part-to-whole relationship with the hierarchical structure of the host-stress interactions of the specific stress. In addition, in order to alleviate over-fitting, a photochemical vegetation indices-based filter is placed as pre-processing operator to remove the non-photochemical noises from the input Sentinel-2 time series.
A Biologically Interpretable Two-stage Deep Neural Network (BIT-DNN) For Hyperspectral Imagery Classification
Apr 19, 2020

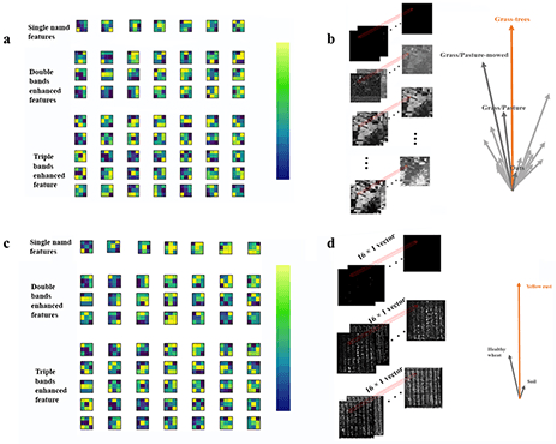
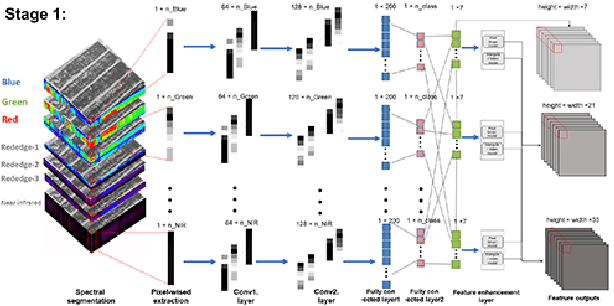
Abstract:Spectral-spatial based deep learning models have recently proven to be effective in hyperspectral image (HSI) classification for various earth monitoring applications such as land cover classification and agricultural monitoring. However, due to the nature of "black-box" model representation, how to explain and interpret the learning process and the model decision remains an open problem. This study proposes an interpretable deep learning model -- a biologically interpretable two-stage deep neural network (BIT-DNN), by integrating biochemical and biophysical associated information into the proposed framework, capable of achieving both high accuracy and interpretability on HSI based classification tasks. The proposed model introduces a two-stage feature learning process. In the first stage, an enhanced interpretable feature block extracts low-level spectral features associated with the biophysical and biochemical attributes of the target entities; and in the second stage, an interpretable capsule block extracts and encapsulates the high-level joint spectral-spatial features into the featured tensors representing the hierarchical structure of the biophysical and biochemical attributes of the target ground entities, which provides the model an improved performance on classification and intrinsic interpretability. We have tested and evaluated the model using two real HSI datasets for crop type recognition and crop disease recognition tasks and compared it with six state-of-the-art machine learning models. The results demonstrate that the proposed model has competitive advantages in terms of both classification accuracy and model interpretability.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge