Tongle Hu
A Latent Encoder Coupled Generative Adversarial Network (LE-GAN) for Efficient Hyperspectral Image Super-resolution
Nov 16, 2021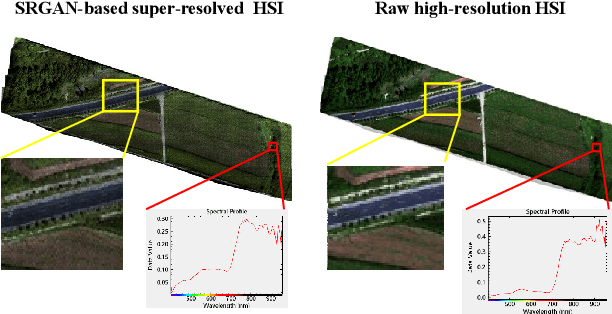
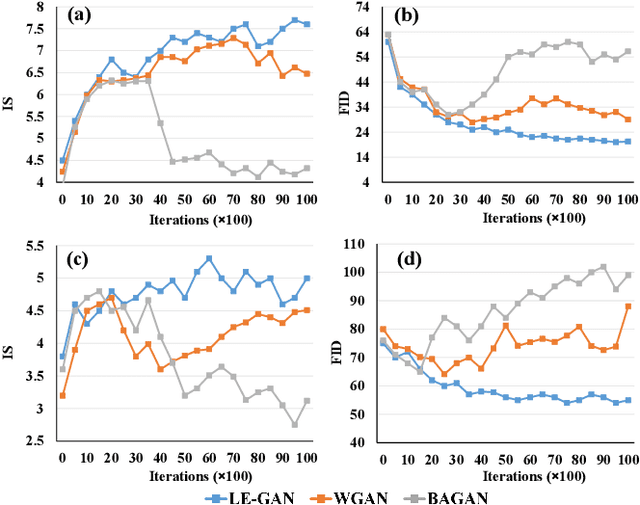
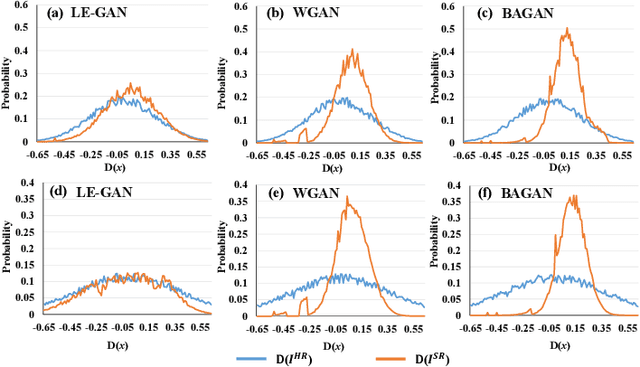
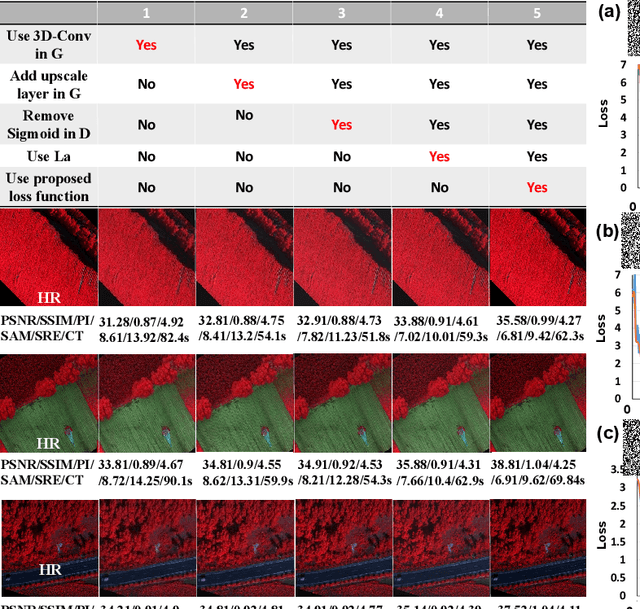
Abstract:Realistic hyperspectral image (HSI) super-resolution (SR) techniques aim to generate a high-resolution (HR) HSI with higher spectral and spatial fidelity from its low-resolution (LR) counterpart. The generative adversarial network (GAN) has proven to be an effective deep learning framework for image super-resolution. However, the optimisation process of existing GAN-based models frequently suffers from the problem of mode collapse, leading to the limited capacity of spectral-spatial invariant reconstruction. This may cause the spectral-spatial distortion on the generated HSI, especially with a large upscaling factor. To alleviate the problem of mode collapse, this work has proposed a novel GAN model coupled with a latent encoder (LE-GAN), which can map the generated spectral-spatial features from the image space to the latent space and produce a coupling component to regularise the generated samples. Essentially, we treat an HSI as a high-dimensional manifold embedded in a latent space. Thus, the optimisation of GAN models is converted to the problem of learning the distributions of high-resolution HSI samples in the latent space, making the distributions of the generated super-resolution HSIs closer to those of their original high-resolution counterparts. We have conducted experimental evaluations on the model performance of super-resolution and its capability in alleviating mode collapse. The proposed approach has been tested and validated based on two real HSI datasets with different sensors (i.e. AVIRIS and UHD-185) for various upscaling factors and added noise levels, and compared with the state-of-the-art super-resolution models (i.e. HyCoNet, LTTR, BAGAN, SR- GAN, WGAN).
A Novel CropdocNet for Automated Potato Late Blight Disease Detection from the Unmanned Aerial Vehicle-based Hyperspectral Imagery
Jul 28, 2021



Abstract:Late blight disease is one of the most destructive diseases in potato crop, leading to serious yield losses globally. Accurate diagnosis of the disease at early stage is critical for precision disease control and management. Current farm practices in crop disease diagnosis are based on manual visual inspection, which is costly, time consuming, subject to individual bias. Recent advances in imaging sensors (e.g. RGB, multiple spectral and hyperspectral cameras), remote sensing and machine learning offer the opportunity to address this challenge. Particularly, hyperspectral imagery (HSI) combining with machine learning/deep learning approaches is preferable for accurately identifying specific plant diseases because the HSI consists of a wide range of high-quality reflectance information beyond human vision, capable of capturing both spectral-spatial information. The proposed method considers the potential disease specific reflectance radiation variance caused by the canopy structural diversity, introduces the multiple capsule layers to model the hierarchical structure of the spectral-spatial disease attributes with the encapsulated features to represent the various classes and the rotation invariance of the disease attributes in the feature space. We have evaluated the proposed method with the real UAV-based HSI data under the controlled field conditions. The effectiveness of the hierarchical features has been quantitatively assessed and compared with the existing representative machine learning/deep learning methods. The experiment results show that the proposed model significantly improves the accuracy performance when considering hierarchical-structure of spectral-spatial features, comparing to the existing methods only using spectral, or spatial or spectral-spatial features without consider hierarchical-structure of spectral-spatial features.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge