Colin Zhang
Uni-Instruct: One-step Diffusion Model through Unified Diffusion Divergence Instruction
May 27, 2025Abstract:In this paper, we unify more than 10 existing one-step diffusion distillation approaches, such as Diff-Instruct, DMD, SIM, SiD, $f$-distill, etc, inside a theory-driven framework which we name the \textbf{\emph{Uni-Instruct}}. Uni-Instruct is motivated by our proposed diffusion expansion theory of the $f$-divergence family. Then we introduce key theories that overcome the intractability issue of the original expanded $f$-divergence, resulting in an equivalent yet tractable loss that effectively trains one-step diffusion models by minimizing the expanded $f$-divergence family. The novel unification introduced by Uni-Instruct not only offers new theoretical contributions that help understand existing approaches from a high-level perspective but also leads to state-of-the-art one-step diffusion generation performances. On the CIFAR10 generation benchmark, Uni-Instruct achieves record-breaking Frechet Inception Distance (FID) values of \textbf{\emph{1.46}} for unconditional generation and \textbf{\emph{1.38}} for conditional generation. On the ImageNet-$64\times 64$ generation benchmark, Uni-Instruct achieves a new SoTA one-step generation FID of \textbf{\emph{1.02}}, which outperforms its 79-step teacher diffusion with a significant improvement margin of 1.33 (1.02 vs 2.35). We also apply Uni-Instruct on broader tasks like text-to-3D generation. For text-to-3D generation, Uni-Instruct gives decent results, which slightly outperforms previous methods, such as SDS and VSD, in terms of both generation quality and diversity. Both the solid theoretical and empirical contributions of Uni-Instruct will potentially help future studies on one-step diffusion distillation and knowledge transferring of diffusion models.
Diff-Instruct*: Towards Human-Preferred One-step Text-to-image Generative Models
Oct 28, 2024
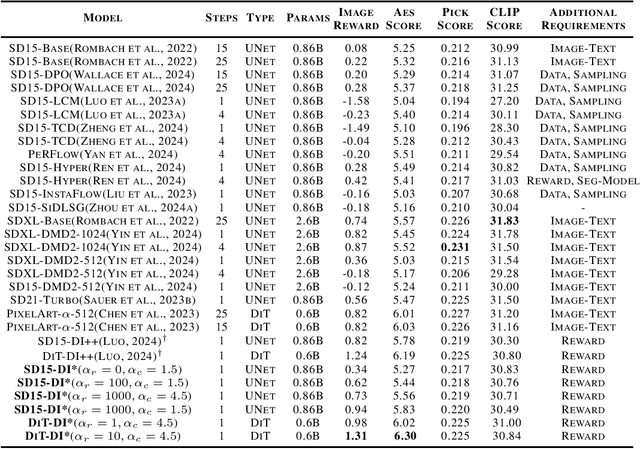

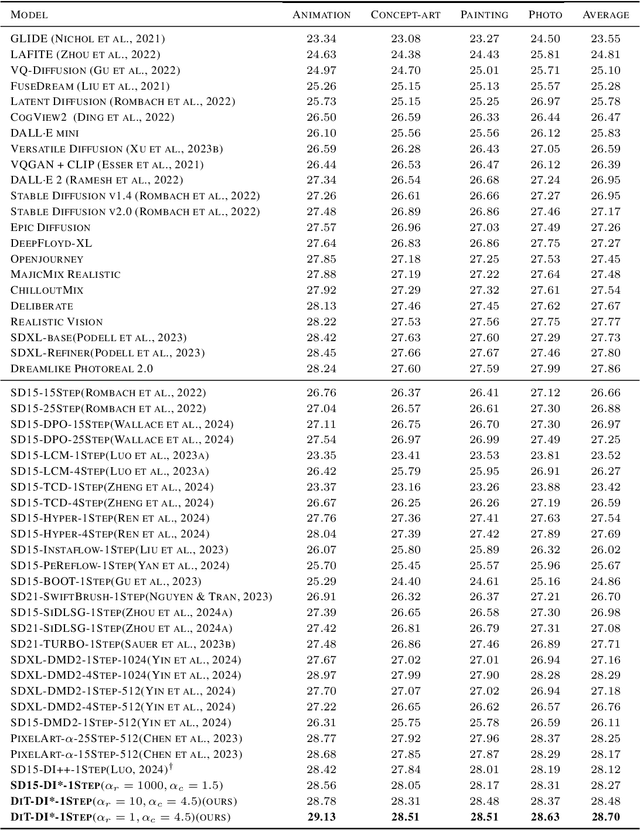
Abstract:In this paper, we introduce the Diff-Instruct*(DI*), a data-free approach for building one-step text-to-image generative models that align with human preference while maintaining the ability to generate highly realistic images. We frame human preference alignment as online reinforcement learning using human feedback (RLHF), where the goal is to maximize the reward function while regularizing the generator distribution to remain close to a reference diffusion process. Unlike traditional RLHF approaches, which rely on the KL divergence for regularization, we introduce a novel score-based divergence regularization, which leads to significantly better performances. Although the direct calculation of this divergence remains intractable, we demonstrate that we can efficiently compute its \emph{gradient} by deriving an equivalent yet tractable loss function. Remarkably, with Stable Diffusion V1.5 as the reference diffusion model, DI* outperforms \emph{all} previously leading models by a large margin. When using the 0.6B PixelArt-$\alpha$ model as the reference diffusion, DI* achieves a new record Aesthetic Score of 6.30 and an Image Reward of 1.31 with only a single generation step, almost doubling the scores of the rest of the models with similar sizes. It also achieves an HPSv2 score of 28.70, establishing a new state-of-the-art benchmark. We also observe that DI* can improve the layout and enrich the colors of generated images.
Untie the Knots: An Efficient Data Augmentation Strategy for Long-Context Pre-Training in Language Models
Sep 07, 2024Abstract:Large language models (LLM) have prioritized expanding the context window from which models can incorporate more information. However, training models to handle long contexts presents significant challenges. These include the scarcity of high-quality natural long-context data, the potential for performance degradation on short-context tasks, and the reduced training efficiency associated with attention mechanisms. In this paper, we introduce Untie the Knots (\textbf{UtK}), a novel data augmentation strategy employed during the continue pre-training phase, designed to efficiently enable LLMs to gain long-context capabilities without the need to modify the existing data mixture. In particular, we chunk the documents, shuffle the chunks, and create a complex and knotted structure of long texts; LLMs are then trained to untie these knots and identify relevant segments within seemingly chaotic token sequences. This approach greatly improves the model's performance by accurately attending to relevant information in long context and the training efficiency is also largely increased. We conduct extensive experiments on models with 7B and 72B parameters, trained on 20 billion tokens, demonstrating that UtK achieves 75\% and 84.5\% accurracy on RULER at 128K context length, significantly outperforming other long context strategies. The trained models will open-source for further research.
Predicting Drug Solubility Using Different Machine Learning Methods -- Linear Regression Model with Extracted Chemical Features vs Graph Convolutional Neural Network
Aug 23, 2023



Abstract:Predicting the solubility of given molecules is an important task in the pharmaceutical industry, and consequently this is a well-studied topic. In this research, we revisited this problem with the advantage of modern computing resources. We applied two machine learning models, a linear regression model and a graph convolutional neural network model, on multiple experimental datasets. Both methods can make reasonable predictions while the GCNN model had the best performance. However, the current GCNN model is a black box, while feature importance analysis from the linear regression model offers more insights into the underlying chemical influences. Using the linear regression model, we show how each functional group affects the overall solubility. Ultimately, knowing how chemical structure influences chemical properties is crucial when designing new drugs. Future work should aim to combine the high performance of GCNNs with the interpretability of linear regression, unlocking new advances in next generation high throughput screening.
Developing novel ligands with enhanced binding affinity for the sphingosine 1-phosphate receptor 1 using machine learning
Jul 29, 2023Abstract:Multiple sclerosis (MS) is a debilitating neurological disease affecting nearly one million people in the United States. Sphingosine-1-phosphate receptor 1, or S1PR1, is a protein target for MS. Siponimod, a ligand of S1PR1, was approved by the FDA in 2019 for MS treatment, but there is a demonstrated need for better therapies. To this end, we finetuned an autoencoder machine learning model that converts chemical formulas into mathematical vectors and generated over 500 molecular variants based on siponimod, out of which 25 compounds had higher predicted binding affinity to S1PR1. The model was able to generate these ligands in just under one hour. Filtering these compounds led to the discovery of six promising candidates with good drug-like properties and ease of synthesis. Furthermore, by analyzing the binding interactions for these ligands, we uncovered several chemical properties that contribute to high binding affinity to S1PR1. This study demonstrates that machine learning can accelerate the drug discovery process and reveal new insights into protein-drug interactions.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge