Cicero Dos Santos
Sobolev Independence Criterion
Oct 31, 2019



Abstract:We propose the Sobolev Independence Criterion (SIC), an interpretable dependency measure between a high dimensional random variable X and a response variable Y . SIC decomposes to the sum of feature importance scores and hence can be used for nonlinear feature selection. SIC can be seen as a gradient regularized Integral Probability Metric (IPM) between the joint distribution of the two random variables and the product of their marginals. We use sparsity inducing gradient penalties to promote input sparsity of the critic of the IPM. In the kernel version we show that SIC can be cast as a convex optimization problem by introducing auxiliary variables that play an important role in feature selection as they are normalized feature importance scores. We then present a neural version of SIC where the critic is parameterized as a homogeneous neural network, improving its representation power as well as its interpretability. We conduct experiments validating SIC for feature selection in synthetic and real-world experiments. We show that SIC enables reliable and interpretable discoveries, when used in conjunction with the holdout randomization test and knockoffs to control the False Discovery Rate. Code is available at http://github.com/ibm/sic.
Wasserstein Barycenter Model Ensembling
Feb 13, 2019



Abstract:In this paper we propose to perform model ensembling in a multiclass or a multilabel learning setting using Wasserstein (W.) barycenters. Optimal transport metrics, such as the Wasserstein distance, allow incorporating semantic side information such as word embeddings. Using W. barycenters to find the consensus between models allows us to balance confidence and semantics in finding the agreement between the models. We show applications of Wasserstein ensembling in attribute-based classification, multilabel learning and image captioning generation. These results show that the W. ensembling is a viable alternative to the basic geometric or arithmetic mean ensembling.
PepCVAE: Semi-Supervised Targeted Design of Antimicrobial Peptide Sequences
Oct 22, 2018
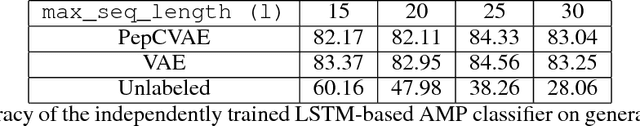
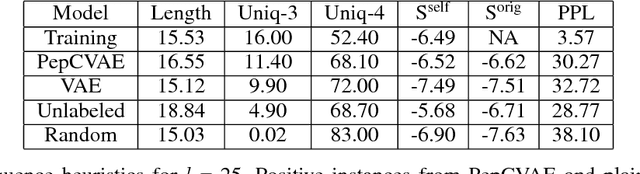
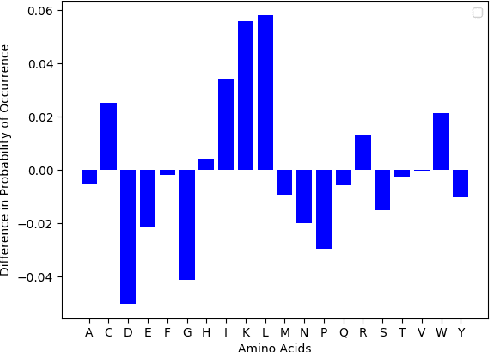
Abstract:Given the emerging global threat of antimicrobial resistance, new methods for next-generation antimicrobial design are urgently needed. We report a peptide generation framework PepCVAE, based on a semi-supervised variational autoencoder (VAE) model, for designing novel antimicrobial peptide (AMP) sequences. Our model learns a rich latent space of the biological peptide context by taking advantage of abundant, unlabeled peptide sequences. The model further learns a disentangled antimicrobial attribute space by using the feedback from a jointly trained AMP classifier that uses limited labeled instances. The disentangled representation allows for controllable generation of AMPs. Extensive analysis of the PepCVAE-generated sequences reveals superior performance of our model in comparison to a plain VAE, as PepCVAE generates novel AMP sequences with higher long-range diversity, while being closer to the training distribution of biological peptides. These features are highly desired in next-generation antimicrobial design.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge