Chuou Xu
Weaver: Foundation Models for Creative Writing
Jan 30, 2024

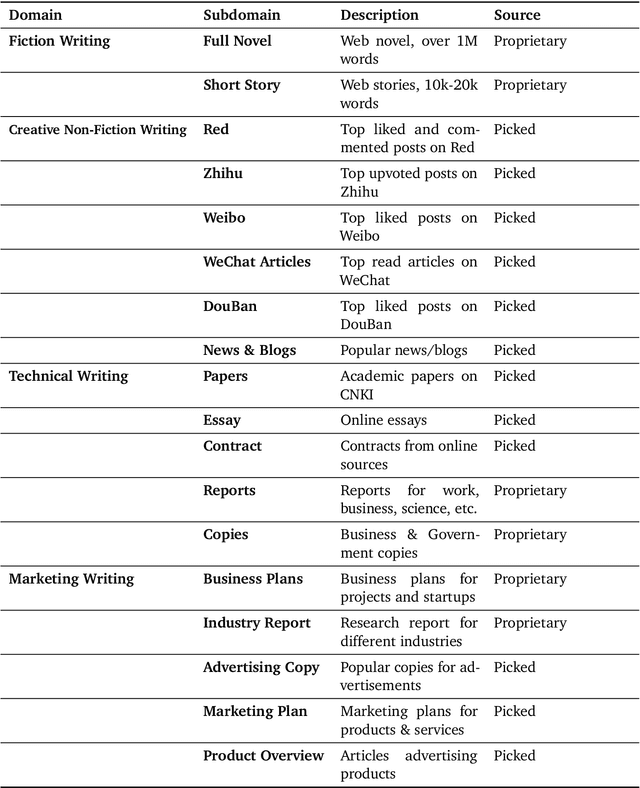

Abstract:This work introduces Weaver, our first family of large language models (LLMs) dedicated to content creation. Weaver is pre-trained on a carefully selected corpus that focuses on improving the writing capabilities of large language models. We then fine-tune Weaver for creative and professional writing purposes and align it to the preference of professional writers using a suit of novel methods for instruction data synthesis and LLM alignment, making it able to produce more human-like texts and follow more diverse instructions for content creation. The Weaver family consists of models of Weaver Mini (1.8B), Weaver Base (6B), Weaver Pro (14B), and Weaver Ultra (34B) sizes, suitable for different applications and can be dynamically dispatched by a routing agent according to query complexity to balance response quality and computation cost. Evaluation on a carefully curated benchmark for assessing the writing capabilities of LLMs shows Weaver models of all sizes outperform generalist LLMs several times larger than them. Notably, our most-capable Weaver Ultra model surpasses GPT-4, a state-of-the-art generalist LLM, on various writing scenarios, demonstrating the advantage of training specialized LLMs for writing purposes. Moreover, Weaver natively supports retrieval-augmented generation (RAG) and function calling (tool usage). We present various use cases of these abilities for improving AI-assisted writing systems, including integration of external knowledge bases, tools, or APIs, and providing personalized writing assistance. Furthermore, we discuss and summarize a guideline and best practices for pre-training and fine-tuning domain-specific LLMs.
Advancing COVID-19 Diagnosis with Privacy-Preserving Collaboration in Artificial Intelligence
Nov 18, 2021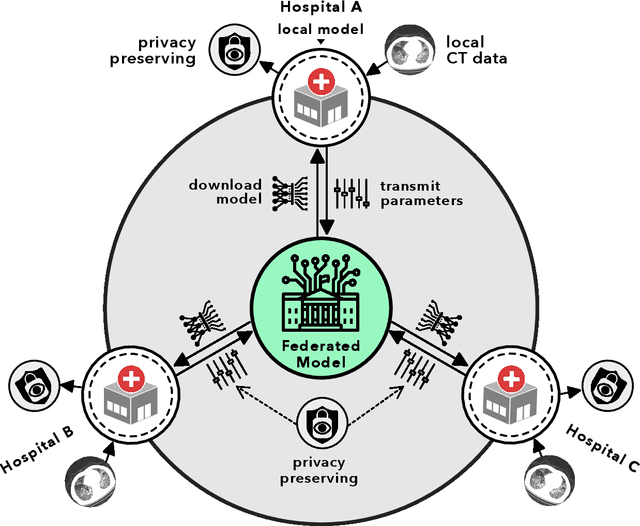
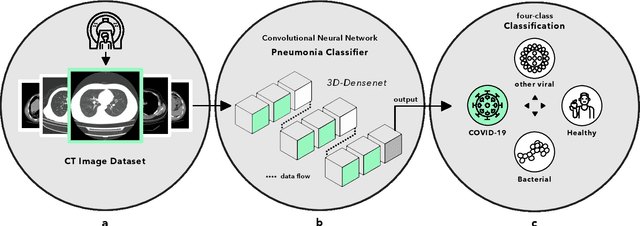
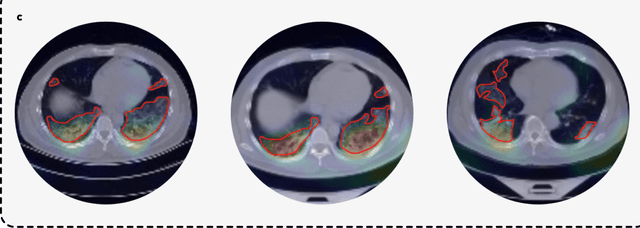
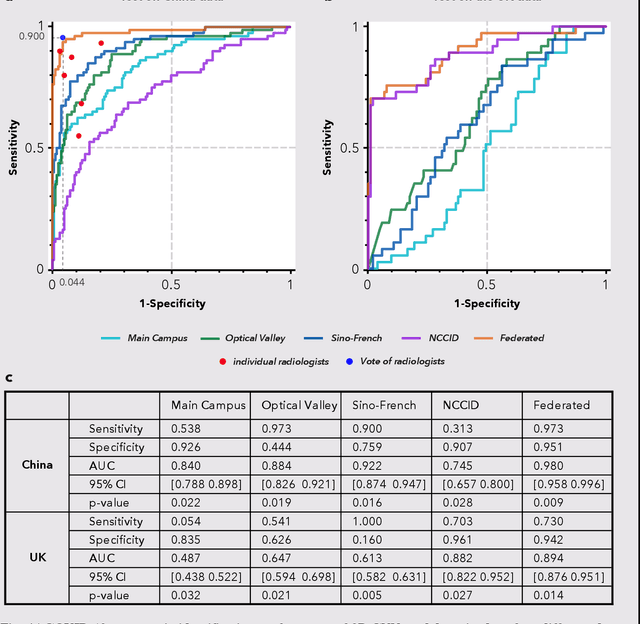
Abstract:Artificial intelligence (AI) provides a promising substitution for streamlining COVID-19 diagnoses. However, concerns surrounding security and trustworthiness impede the collection of large-scale representative medical data, posing a considerable challenge for training a well-generalised model in clinical practices. To address this, we launch the Unified CT-COVID AI Diagnostic Initiative (UCADI), where the AI model can be distributedly trained and independently executed at each host institution under a federated learning framework (FL) without data sharing. Here we show that our FL model outperformed all the local models by a large yield (test sensitivity /specificity in China: 0.973/0.951, in the UK: 0.730/0.942), achieving comparable performance with a panel of professional radiologists. We further evaluated the model on the hold-out (collected from another two hospitals leaving out the FL) and heterogeneous (acquired with contrast materials) data, provided visual explanations for decisions made by the model, and analysed the trade-offs between the model performance and the communication costs in the federated training process. Our study is based on 9,573 chest computed tomography scans (CTs) from 3,336 patients collected from 23 hospitals located in China and the UK. Collectively, our work advanced the prospects of utilising federated learning for privacy-preserving AI in digital health.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge