Chuansheng Zheng
A DeepSeek-Powered AI System for Automated Chest Radiograph Interpretation in Clinical Practice
Dec 23, 2025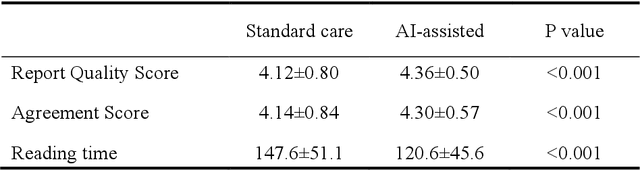
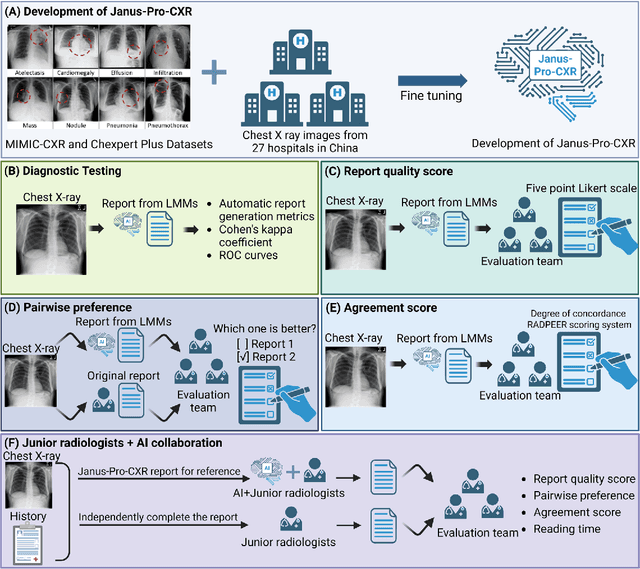
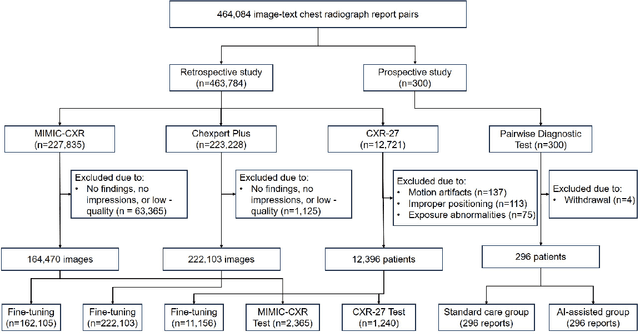
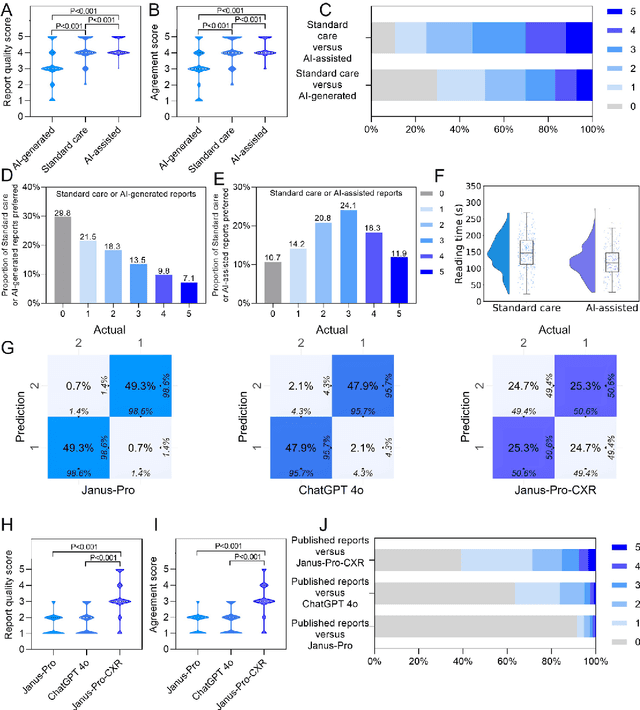
Abstract:A global shortage of radiologists has been exacerbated by the significant volume of chest X-ray workloads, particularly in primary care. Although multimodal large language models show promise, existing evaluations predominantly rely on automated metrics or retrospective analyses, lacking rigorous prospective clinical validation. Janus-Pro-CXR (1B), a chest X-ray interpretation system based on DeepSeek Janus-Pro model, was developed and rigorously validated through a multicenter prospective trial (NCT07117266). Our system outperforms state-of-the-art X-ray report generation models in automated report generation, surpassing even larger-scale models including ChatGPT 4o (200B parameters), while demonstrating reliable detection of six clinically critical radiographic findings. Retrospective evaluation confirms significantly higher report accuracy than Janus-Pro and ChatGPT 4o. In prospective clinical deployment, AI assistance significantly improved report quality scores, reduced interpretation time by 18.3% (P < 0.001), and was preferred by a majority of experts in 54.3% of cases. Through lightweight architecture and domain-specific optimization, Janus-Pro-CXR improves diagnostic reliability and workflow efficiency, particularly in resource-constrained settings. The model architecture and implementation framework will be open-sourced to facilitate the clinical translation of AI-assisted radiology solutions.
MoSt-DSA: Modeling Motion and Structural Interactions for Direct Multi-Frame Interpolation in DSA Images
Jul 09, 2024



Abstract:Artificial intelligence has become a crucial tool for medical image analysis. As an advanced cerebral angiography technique, Digital Subtraction Angiography (DSA) poses a challenge where the radiation dose to humans is proportional to the image count. By reducing images and using AI interpolation instead, the radiation can be cut significantly. However, DSA images present more complex motion and structural features than natural scenes, making interpolation more challenging. We propose MoSt-DSA, the first work that uses deep learning for DSA frame interpolation. Unlike natural scene Video Frame Interpolation (VFI) methods that extract unclear or coarse-grained features, we devise a general module that models motion and structural context interactions between frames in an efficient full convolution manner by adjusting optimal context range and transforming contexts into linear functions. Benefiting from this, MoSt-DSA is also the first method that directly achieves any number of interpolations at any time steps with just one forward pass during both training and testing. We conduct extensive comparisons with 7 representative VFI models for interpolating 1 to 3 frames, MoSt-DSA demonstrates robust results across 470 DSA image sequences (each typically 152 images), with average SSIM over 0.93, average PSNR over 38 (standard deviations of less than 0.030 and 3.6, respectively), comprehensively achieving state-of-the-art performance in accuracy, speed, visual effect, and memory usage. Our code is available at https://github.com/ZyoungXu/MoSt-DSA.
3D Vessel Reconstruction from Sparse-View Dynamic DSA Images via Vessel Probability Guided Attenuation Learning
May 17, 2024



Abstract:Digital Subtraction Angiography (DSA) is one of the gold standards in vascular disease diagnosing. With the help of contrast agent, time-resolved 2D DSA images deliver comprehensive insights into blood flow information and can be utilized to reconstruct 3D vessel structures. Current commercial DSA systems typically demand hundreds of scanning views to perform reconstruction, resulting in substantial radiation exposure. However, sparse-view DSA reconstruction, aimed at reducing radiation dosage, is still underexplored in the research community. The dynamic blood flow and insufficient input of sparse-view DSA images present significant challenges to the 3D vessel reconstruction task. In this study, we propose to use a time-agnostic vessel probability field to solve this problem effectively. Our approach, termed as vessel probability guided attenuation learning, represents the DSA imaging as a complementary weighted combination of static and dynamic attenuation fields, with the weights derived from the vessel probability field. Functioning as a dynamic mask, vessel probability provides proper gradients for both static and dynamic fields adaptive to different scene types. This mechanism facilitates a self-supervised decomposition between static backgrounds and dynamic contrast agent flow, and significantly improves the reconstruction quality. Our model is trained by minimizing the disparity between synthesized projections and real captured DSA images. We further employ two training strategies to improve our reconstruction quality: (1) coarse-to-fine progressive training to achieve better geometry and (2) temporal perturbed rendering loss to enforce temporal consistency. Experimental results have demonstrated superior quality on both 3D vessel reconstruction and 2D view synthesis.
TOGS: Gaussian Splatting with Temporal Opacity Offset for Real-Time 4D DSA Rendering
Mar 28, 2024Abstract:Four-dimensional Digital Subtraction Angiography (4D DSA) is a medical imaging technique that provides a series of 2D images captured at different stages and angles during the process of contrast agent filling blood vessels. It plays a significant role in the diagnosis of cerebrovascular diseases. Improving the rendering quality and speed under sparse sampling is important for observing the status and location of lesions. The current methods exhibit inadequate rendering quality in sparse views and suffer from slow rendering speed. To overcome these limitations, we propose TOGS, a Gaussian splatting method with opacity offset over time, which can effectively improve the rendering quality and speed of 4D DSA. We introduce an opacity offset table for each Gaussian to model the temporal variations in the radiance of the contrast agent. By interpolating the opacity offset table, the opacity variation of the Gaussian at different time points can be determined. This enables us to render the 2D DSA image at that specific moment. Additionally, we introduced a Smooth loss term in the loss function to mitigate overfitting issues that may arise in the model when dealing with sparse view scenarios. During the training phase, we randomly prune Gaussians, thereby reducing the storage overhead of the model. The experimental results demonstrate that compared to previous methods, this model achieves state-of-the-art reconstruction quality under the same number of training views. Additionally, it enables real-time rendering while maintaining low storage overhead. The code will be publicly available.
ConvFormer: Plug-and-Play CNN-Style Transformers for Improving Medical Image Segmentation
Sep 09, 2023Abstract:Transformers have been extensively studied in medical image segmentation to build pairwise long-range dependence. Yet, relatively limited well-annotated medical image data makes transformers struggle to extract diverse global features, resulting in attention collapse where attention maps become similar or even identical. Comparatively, convolutional neural networks (CNNs) have better convergence properties on small-scale training data but suffer from limited receptive fields. Existing works are dedicated to exploring the combinations of CNN and transformers while ignoring attention collapse, leaving the potential of transformers under-explored. In this paper, we propose to build CNN-style Transformers (ConvFormer) to promote better attention convergence and thus better segmentation performance. Specifically, ConvFormer consists of pooling, CNN-style self-attention (CSA), and convolutional feed-forward network (CFFN) corresponding to tokenization, self-attention, and feed-forward network in vanilla vision transformers. In contrast to positional embedding and tokenization, ConvFormer adopts 2D convolution and max-pooling for both position information preservation and feature size reduction. In this way, CSA takes 2D feature maps as inputs and establishes long-range dependency by constructing self-attention matrices as convolution kernels with adaptive sizes. Following CSA, 2D convolution is utilized for feature refinement through CFFN. Experimental results on multiple datasets demonstrate the effectiveness of ConvFormer working as a plug-and-play module for consistent performance improvement of transformer-based frameworks. Code is available at https://github.com/xianlin7/ConvFormer.
TiAVox: Time-aware Attenuation Voxels for Sparse-view 4D DSA Reconstruction
Sep 05, 2023Abstract:Four-dimensional Digital Subtraction Angiography (4D DSA) plays a critical role in the diagnosis of many medical diseases, such as Arteriovenous Malformations (AVM) and Arteriovenous Fistulas (AVF). Despite its significant application value, the reconstruction of 4D DSA demands numerous views to effectively model the intricate vessels and radiocontrast flow, thereby implying a significant radiation dose. To address this high radiation issue, we propose a Time-aware Attenuation Voxel (TiAVox) approach for sparse-view 4D DSA reconstruction, which paves the way for high-quality 4D imaging. Additionally, 2D and 3D DSA imaging results can be generated from the reconstructed 4D DSA images. TiAVox introduces 4D attenuation voxel grids, which reflect attenuation properties from both spatial and temporal dimensions. It is optimized by minimizing discrepancies between the rendered images and sparse 2D DSA images. Without any neural network involved, TiAVox enjoys specific physical interpretability. The parameters of each learnable voxel represent the attenuation coefficients. We validated the TiAVox approach on both clinical and simulated datasets, achieving a 31.23 Peak Signal-to-Noise Ratio (PSNR) for novel view synthesis using only 30 views on the clinically sourced dataset, whereas traditional Feldkamp-Davis-Kress methods required 133 views. Similarly, with merely 10 views from the synthetic dataset, TiAVox yielded a PSNR of 34.32 for novel view synthesis and 41.40 for 3D reconstruction. We also executed ablation studies to corroborate the essential components of TiAVox. The code will be publically available.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge