Christopher Nimsky
Vertebral body segmentation with GrowCut: Initial experience, workflow and practical application
Nov 13, 2017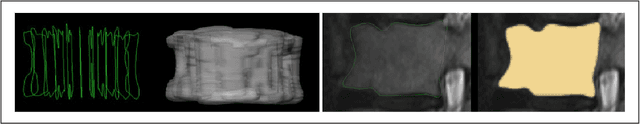
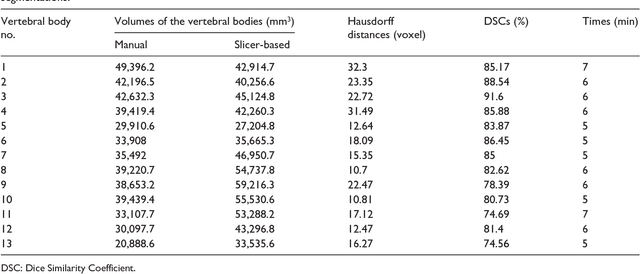
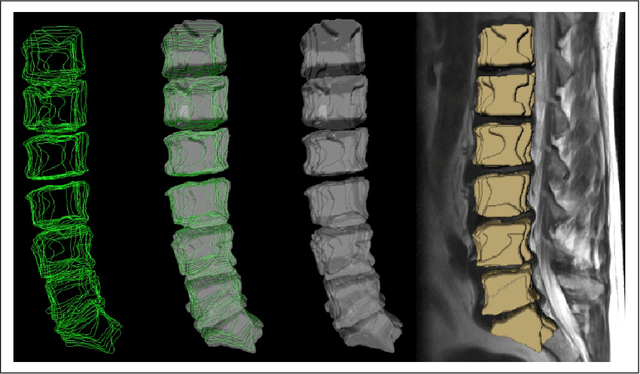

Abstract:In this contribution, we used the GrowCut segmentation algorithm publicly available in three-dimensional Slicer for three-dimensional segmentation of vertebral bodies. To the best of our knowledge, this is the first time that the GrowCut method has been studied for the usage of vertebral body segmentation. In brief, we found that the GrowCut segmentation times were consistently less than the manual segmentation times. Hence, GrowCut provides an alternative to a manual slice-by-slice segmentation process.
* 10 pages
Cellular Automata Segmentation of the Boundary between the Compacta of Vertebral Bodies and Surrounding Structures
Mar 03, 2016Abstract:Due to the aging population, spinal diseases get more and more common nowadays; e.g., lifetime risk of osteoporotic fracture is 40% for white women and 13% for white men in the United States. Thus the numbers of surgical spinal procedures are also increasing with the aging population and precise diagnosis plays a vital role in reducing complication and recurrence of symptoms. Spinal imaging of vertebral column is a tedious process subjected to interpretation errors. In this contribution, we aim to reduce time and error for vertebral interpretation by applying and studying the GrowCut-algorithm for boundary segmentation between vertebral body compacta and surrounding structures. GrowCut is a competitive region growing algorithm using cellular automata. For our study, vertebral T2-weighted Magnetic Resonance Imaging (MRI) scans were first manually outlined by neurosurgeons. Then, the vertebral bodies were segmented in the medical images by a GrowCut-trained physician using the semi-automated GrowCut-algorithm. Afterwards, results of both segmentation processes were compared using the Dice Similarity Coefficient (DSC) and the Hausdorff Distance (HD) which yielded to a DSC of 82.99+/-5.03% and a HD of 18.91+/-7.2 voxel, respectively. In addition, the times have been measured during the manual and the GrowCut segmentations, showing that a GrowCut-segmentation - with an average time of less than six minutes (5.77+/-0.73) - is significantly shorter than a pure manual outlining.
* 6 pages, 5 figures, 1 table, 42 references
Preoperative Volume Determination for Pituitary Adenoma
Feb 05, 2016Abstract:The most common sellar lesion is the pituitary adenoma, and sellar tumors are approximately 10-15% of all intracranial neoplasms. Manual slice-by-slice segmentation takes quite some time that can be reduced by using the appropriate algorithms. In this contribution, we present a segmentation method for pituitary adenoma. The method is based on an algorithm that we have applied recently to segmenting glioblastoma multiforme. A modification of this scheme is used for adenoma segmentation that is much harder to perform, due to lack of contrast-enhanced boundaries. In our experimental evaluation, neurosurgeons performed manual slice-by-slice segmentation of ten magnetic resonance imaging (MRI) cases. The segmentations were compared to the segmentation results of the proposed method using the Dice Similarity Coefficient (DSC). The average DSC for all datasets was 75.92% +/- 7.24%. A manual segmentation took about four minutes and our algorithm required about one second.
Cube-Cut: Vertebral Body Segmentation in MRI-Data through Cubic-Shaped Divergences
Jul 22, 2014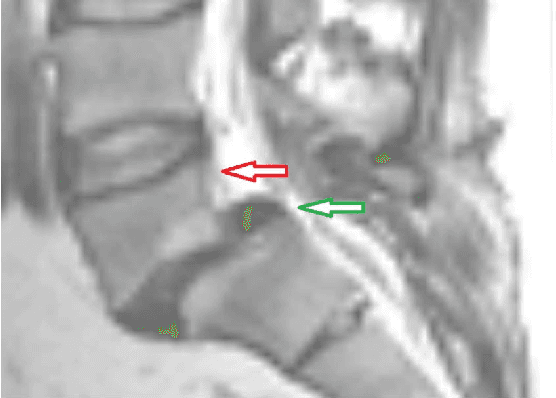
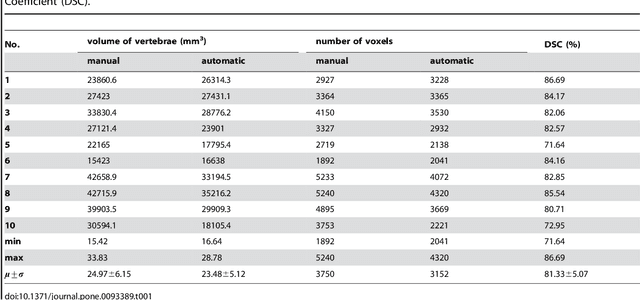
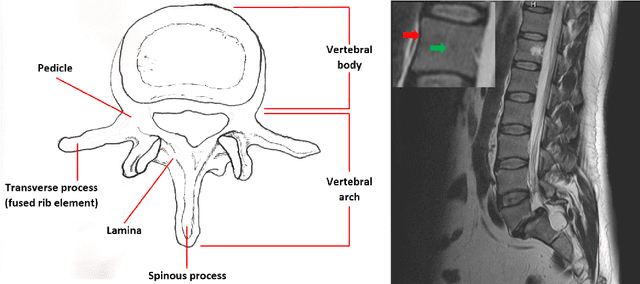
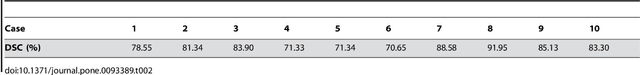
Abstract:In this article, we present a graph-based method using a cubic template for volumetric segmentation of vertebrae in magnetic resonance imaging (MRI) acquisitions. The user can define the degree of deviation from a regular cube via a smoothness value Delta. The Cube-Cut algorithm generates a directed graph with two terminal nodes (s-t-network), where the nodes of the graph correspond to a cubic-shaped subset of the image's voxels. The weightings of the graph's terminal edges, which connect every node with a virtual source s or a virtual sink t, represent the affinity of a voxel to the vertebra (source) and to the background (sink). Furthermore, a set of infinite weighted and non-terminal edges implements the smoothness term. After graph construction, a minimal s-t-cut is calculated within polynomial computation time, which splits the nodes into two disjoint units. Subsequently, the segmentation result is determined out of the source-set. A quantitative evaluation of a C++ implementation of the algorithm resulted in an average Dice Similarity Coefficient (DSC) of 81.33% and a running time of less than a minute.
A Ray-based Approach for Boundary Estimation of Fiber Bundles Derived from Diffusion Tensor Imaging
Oct 23, 2013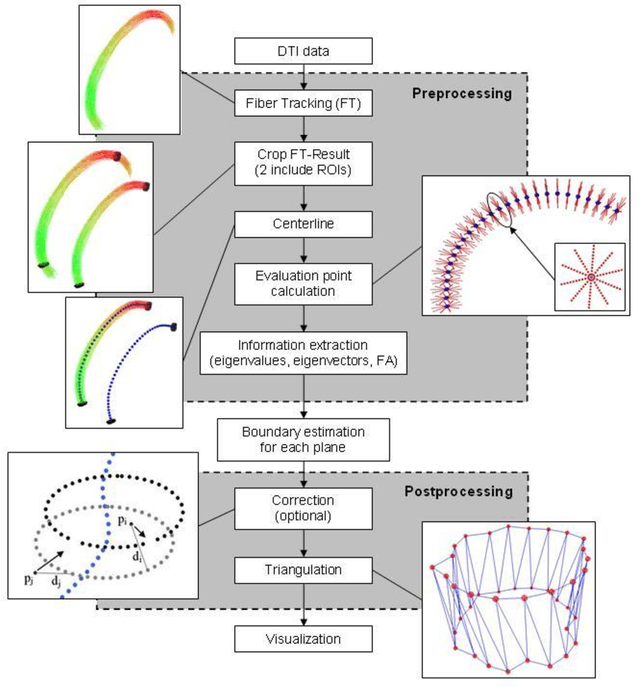
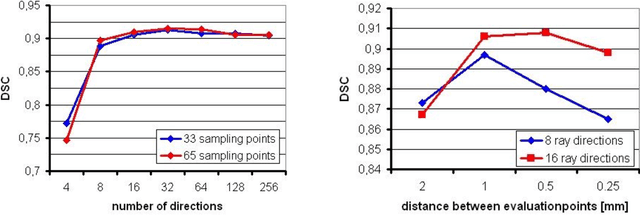
Abstract:Diffusion Tensor Imaging (DTI) is a non-invasive imaging technique that allows estimation of the location of white matter tracts in-vivo, based on the measurement of water diffusion properties. For each voxel, a second-order tensor can be calculated by using diffusion-weighted sequences (DWI) that are sensitive to the random motion of water molecules. Given at least 6 diffusion-weighted images with different gradients and one unweighted image, the coefficients of the symmetric diffusion tensor matrix can be calculated. Deriving the eigensystem of the tensor, the eigenvectors and eigenvalues can be calculated to describe the three main directions of diffusion and its magnitude. Using DTI data, fiber bundles can be determined, to gain information about eloquent brain structures. Especially in neurosurgery, information about location and dimension of eloquent structures like the corticospinal tract or the visual pathways is of major interest. Therefore, the fiber bundle boundary has to be determined. In this paper, a novel ray-based approach for boundary estimation of tubular structures is presented.
* 5 pages, 2 figures, 7 references
Determination, Calculation and Representation of the Upper and Lower Sealing Zones During Virtual Stenting of Aneurysms
Oct 21, 2013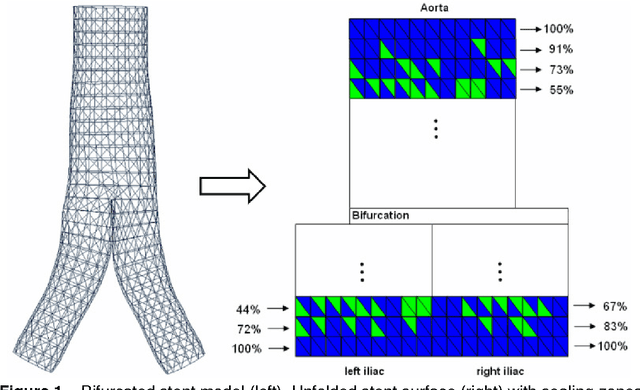
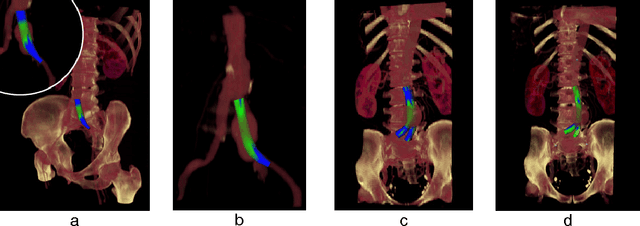
Abstract:In this contribution, a novel method for stent simulation in preoperative computed tomography angiography (CTA) acquisitions of patients is presented where the sealing zones are automatically calculated and visualized. The method is eligible for non-bifurcated and bifurcated stents (Y-stents). Results of the proposed stent simulation with an automatic calculation of the sealing zones for specific diseases (abdominal aortic aneurysms (AAA), thoracic aortic aneurysms (TAA), iliac aneurysms) are presented. The contribution is organized as follows. Section 2 presents the proposed approach. In Section 3, experimental results are discussed. Section 4 concludes the contribution and outlines areas for future work.
* 4 pages, 2 figures, 10 references
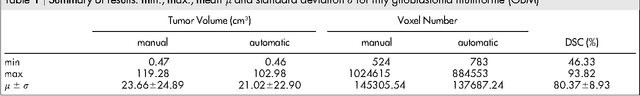
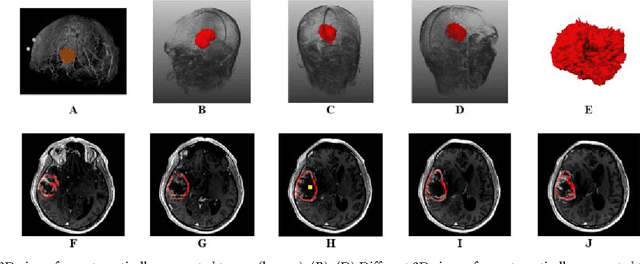
GBM Volumetry using the 3D Slicer Medical Image Computing Platform
Mar 05, 2013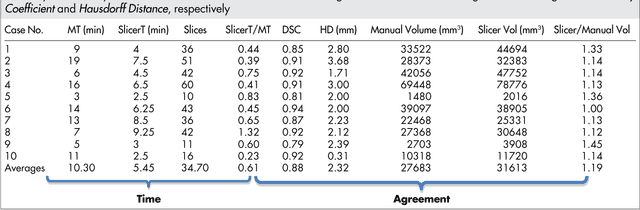
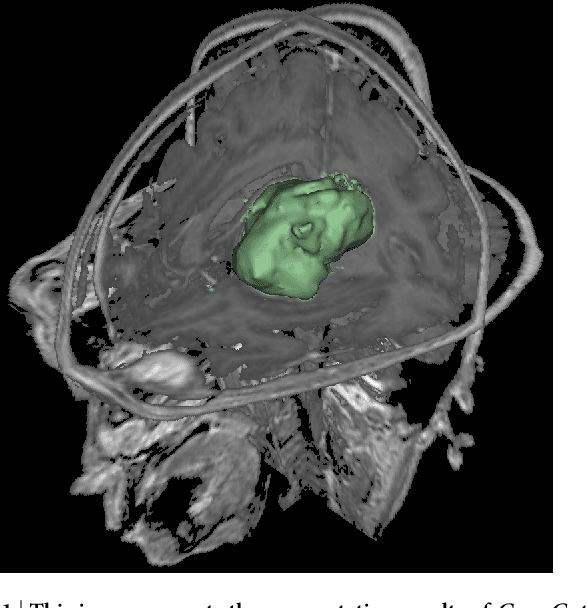
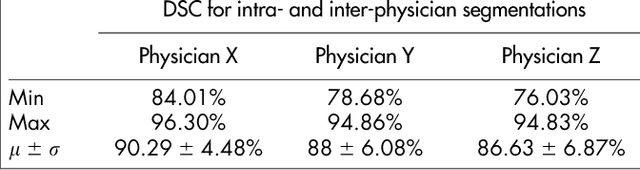
Abstract:Volumetric change in glioblastoma multiforme (GBM) over time is a critical factor in treatment decisions. Typically, the tumor volume is computed on a slice-by-slice basis using MRI scans obtained at regular intervals. (3D)Slicer - a free platform for biomedical research - provides an alternative to this manual slice-by-slice segmentation process, which is significantly faster and requires less user interaction. In this study, 4 physicians segmented GBMs in 10 patients, once using the competitive region-growing based GrowCut segmentation module of Slicer, and once purely by drawing boundaries completely manually on a slice-by-slice basis. Furthermore, we provide a variability analysis for three physicians for 12 GBMs. The time required for GrowCut segmentation was on an average 61% of the time required for a pure manual segmentation. A comparison of Slicer-based segmentation with manual slice-by-slice segmentation resulted in a Dice Similarity Coefficient of 88.43 +/- 5.23% and a Hausdorff Distance of 2.32 +/- 5.23 mm.
* 7 pages, 6 figures, 2 tables, 1 equation, 43 references
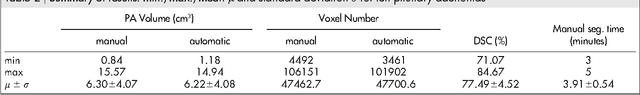
Pituitary Adenoma Volumetry with 3D Slicer
Dec 12, 2012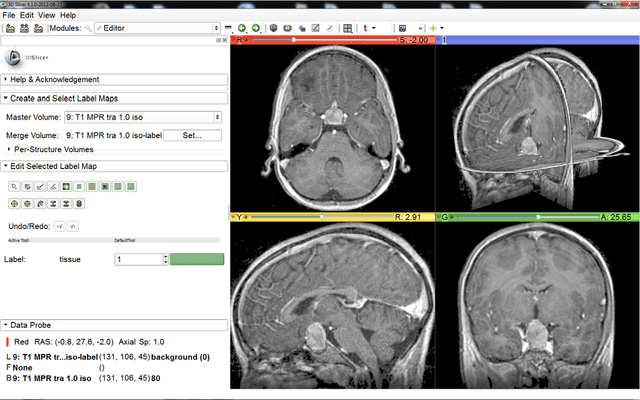
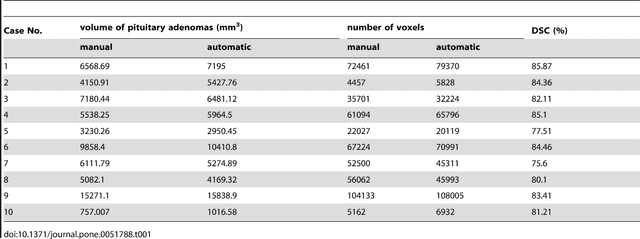
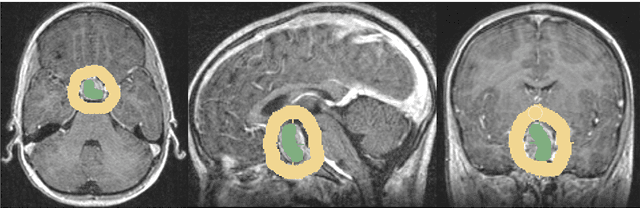
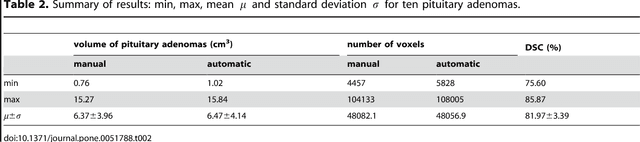
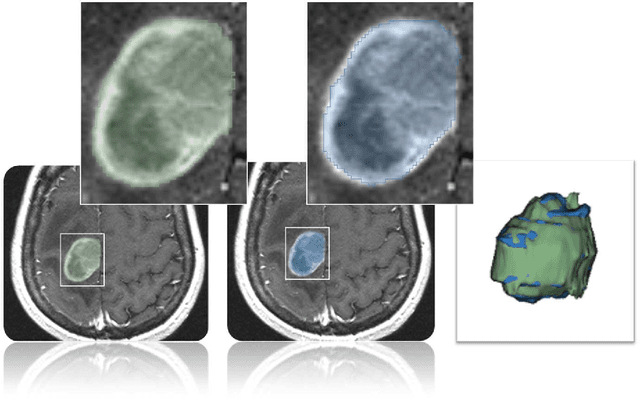
Abstract:In this study, we present pituitary adenoma volumetry using the free and open source medical image computing platform for biomedical research: (3D) Slicer. Volumetric changes in cerebral pathologies like pituitary adenomas are a critical factor in treatment decisions by physicians and in general the volume is acquired manually. Therefore, manual slice-by-slice segmentations in magnetic resonance imaging (MRI) data, which have been obtained at regular intervals, are performed. In contrast to this manual time consuming slice-by-slice segmentation process Slicer is an alternative which can be significantly faster and less user intensive. In this contribution, we compare pure manual segmentations of ten pituitary adenomas with semi-automatic segmentations under Slicer. Thus, physicians drew the boundaries completely manually on a slice-by-slice basis and performed a Slicer-enhanced segmentation using the competitive region-growing based module of Slicer named GrowCut. Results showed that the time and user effort required for GrowCut-based segmentations were on average about thirty percent less than the pure manual segmentations. Furthermore, we calculated the Dice Similarity Coefficient (DSC) between the manual and the Slicer-based segmentations to proof that the two are comparable yielding an average DSC of 81.97\pm3.39%.
* 7 pages, 5 figures, 2 tables, 30 references
Template-Cut: A Pattern-Based Segmentation Paradigm
May 30, 2012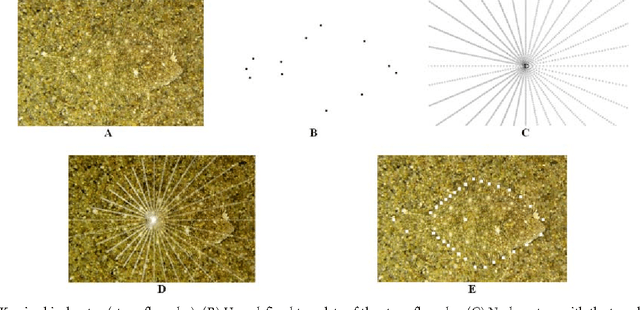
Abstract:We present a scale-invariant, template-based segmentation paradigm that sets up a graph and performs a graph cut to separate an object from the background. Typically graph-based schemes distribute the nodes of the graph uniformly and equidistantly on the image, and use a regularizer to bias the cut towards a particular shape. The strategy of uniform and equidistant nodes does not allow the cut to prefer more complex structures, especially when areas of the object are indistinguishable from the background. We propose a solution by introducing the concept of a "template shape" of the target object in which the nodes are sampled non-uniformly and non-equidistantly on the image. We evaluate it on 2D-images where the object's textures and backgrounds are similar, and large areas of the object have the same gray level appearance as the background. We also evaluate it in 3D on 60 brain tumor datasets for neurosurgical planning purposes.
* 8 pages, 6 figures, 3 tables, 6 equations, 51 references
Square-Cut: A Segmentation Algorithm on the Basis of a Rectangle Shape
Mar 13, 2012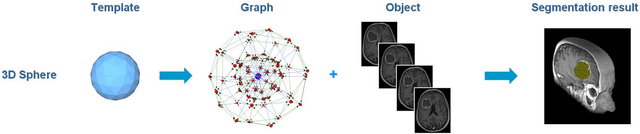
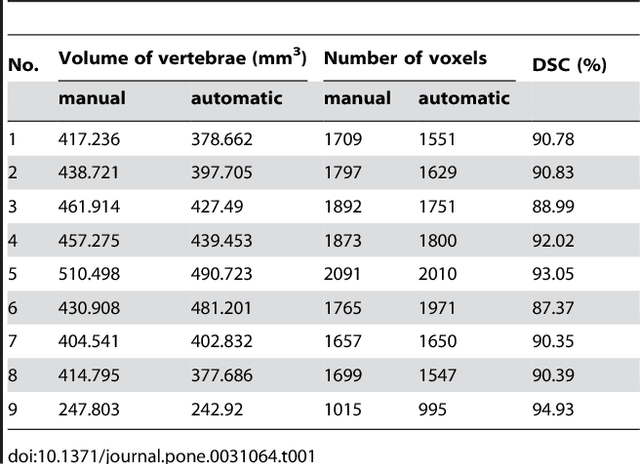
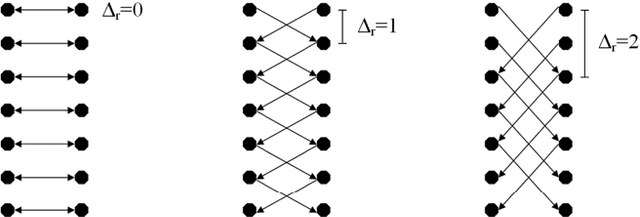
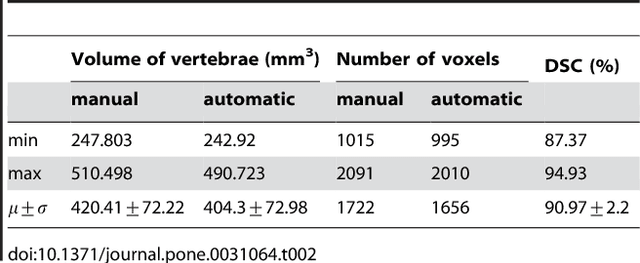
Abstract:We present a rectangle-based segmentation algorithm that sets up a graph and performs a graph cut to separate an object from the background. However, graph-based algorithms distribute the graph's nodes uniformly and equidistantly on the image. Then, a smoothness term is added to force the cut to prefer a particular shape. This strategy does not allow the cut to prefer a certain structure, especially when areas of the object are indistinguishable from the background. We solve this problem by referring to a rectangle shape of the object when sampling the graph nodes, i.e., the nodes are distributed nonuniformly and non-equidistantly on the image. This strategy can be useful, when areas of the object are indistinguishable from the background. For evaluation, we focus on vertebrae images from Magnetic Resonance Imaging (MRI) datasets to support the time consuming manual slice-by-slice segmentation performed by physicians. The ground truth of the vertebrae boundaries were manually extracted by two clinical experts (neurological surgeons) with several years of experience in spine surgery and afterwards compared with the automatic segmentation results of the proposed scheme yielding an average Dice Similarity Coefficient (DSC) of 90.97\pm62.2%.
* 13 pages, 17 figures, 2 tables, 3 equations, 42 references
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge