Daniela Kuhnt
Preoperative Volume Determination for Pituitary Adenoma
Feb 05, 2016Abstract:The most common sellar lesion is the pituitary adenoma, and sellar tumors are approximately 10-15% of all intracranial neoplasms. Manual slice-by-slice segmentation takes quite some time that can be reduced by using the appropriate algorithms. In this contribution, we present a segmentation method for pituitary adenoma. The method is based on an algorithm that we have applied recently to segmenting glioblastoma multiforme. A modification of this scheme is used for adenoma segmentation that is much harder to perform, due to lack of contrast-enhanced boundaries. In our experimental evaluation, neurosurgeons performed manual slice-by-slice segmentation of ten magnetic resonance imaging (MRI) cases. The segmentations were compared to the segmentation results of the proposed method using the Dice Similarity Coefficient (DSC). The average DSC for all datasets was 75.92% +/- 7.24%. A manual segmentation took about four minutes and our algorithm required about one second.
A Ray-based Approach for Boundary Estimation of Fiber Bundles Derived from Diffusion Tensor Imaging
Oct 23, 2013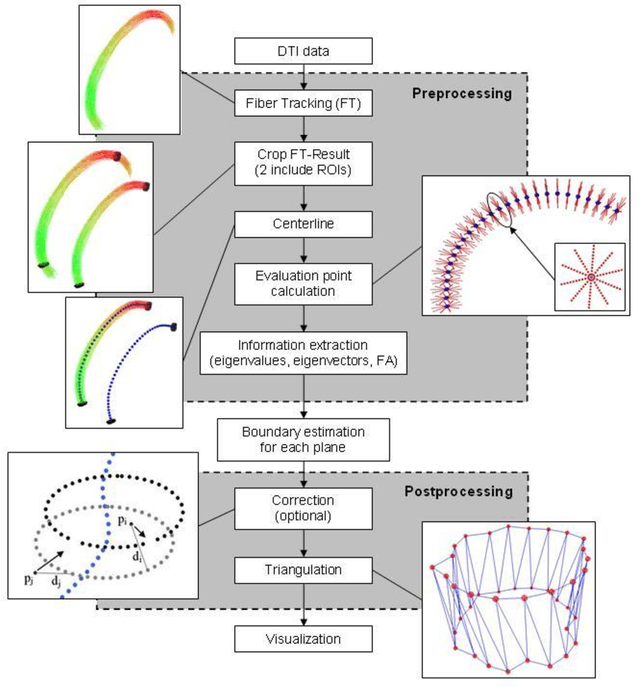
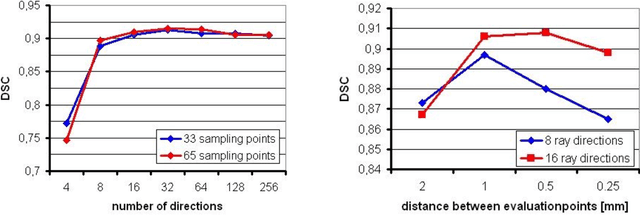
Abstract:Diffusion Tensor Imaging (DTI) is a non-invasive imaging technique that allows estimation of the location of white matter tracts in-vivo, based on the measurement of water diffusion properties. For each voxel, a second-order tensor can be calculated by using diffusion-weighted sequences (DWI) that are sensitive to the random motion of water molecules. Given at least 6 diffusion-weighted images with different gradients and one unweighted image, the coefficients of the symmetric diffusion tensor matrix can be calculated. Deriving the eigensystem of the tensor, the eigenvectors and eigenvalues can be calculated to describe the three main directions of diffusion and its magnitude. Using DTI data, fiber bundles can be determined, to gain information about eloquent brain structures. Especially in neurosurgery, information about location and dimension of eloquent structures like the corticospinal tract or the visual pathways is of major interest. Therefore, the fiber bundle boundary has to be determined. In this paper, a novel ray-based approach for boundary estimation of tubular structures is presented.
* 5 pages, 2 figures, 7 references
A Comparison of Two Human Brain Tumor Segmentation Methods for MRI Data
Mar 10, 2011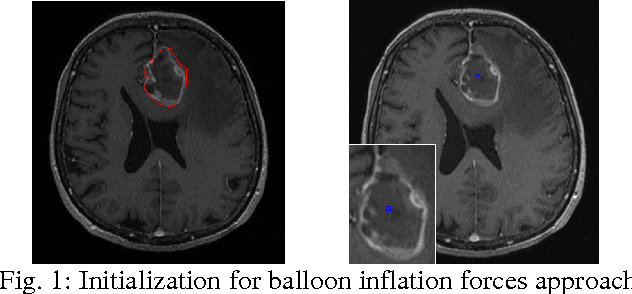
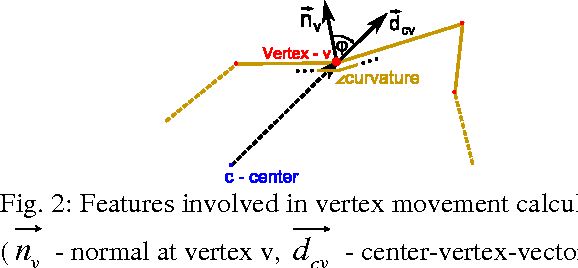
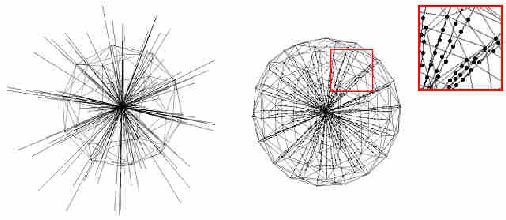
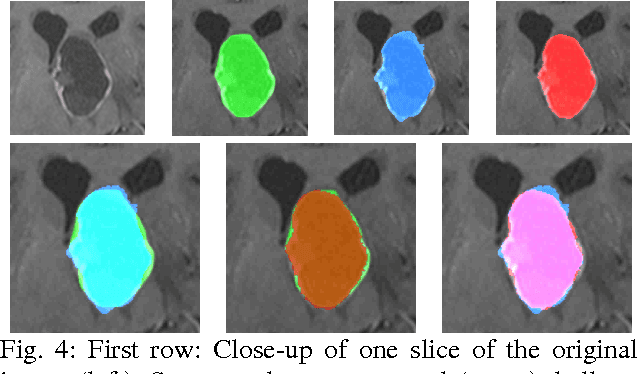
Abstract:The most common primary brain tumors are gliomas, evolving from the cerebral supportive cells. For clinical follow-up, the evaluation of the preoperative tumor volume is essential. Volumetric assessment of tumor volume with manual segmentation of its outlines is a time-consuming process that can be overcome with the help of computerized segmentation methods. In this contribution, two methods for World Health Organization (WHO) grade IV glioma segmentation in the human brain are compared using magnetic resonance imaging (MRI) patient data from the clinical routine. One method uses balloon inflation forces, and relies on detection of high intensity tumor boundaries that are coupled with the use of contrast agent gadolinium. The other method sets up a directed and weighted graph and performs a min-cut for optimal segmentation results. The ground truth of the tumor boundaries - for evaluating the methods on 27 cases - is manually extracted by neurosurgeons with several years of experience in the resection of gliomas. A comparison is performed using the Dice Similarity Coefficient (DSC), a measure for the spatial overlap of different segmentation results.
Ray-Based and Graph-Based Methods for Fiber Bundle Boundary Estimation
Mar 10, 2011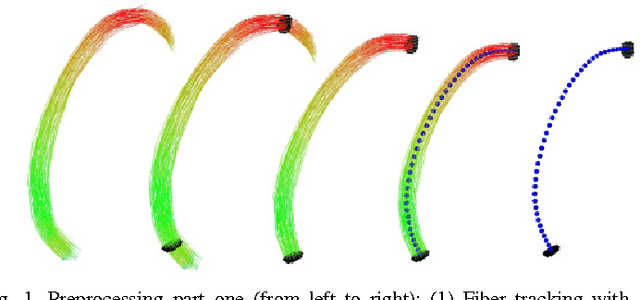
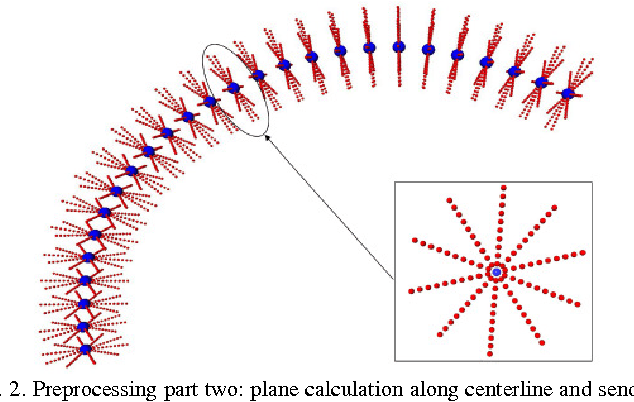
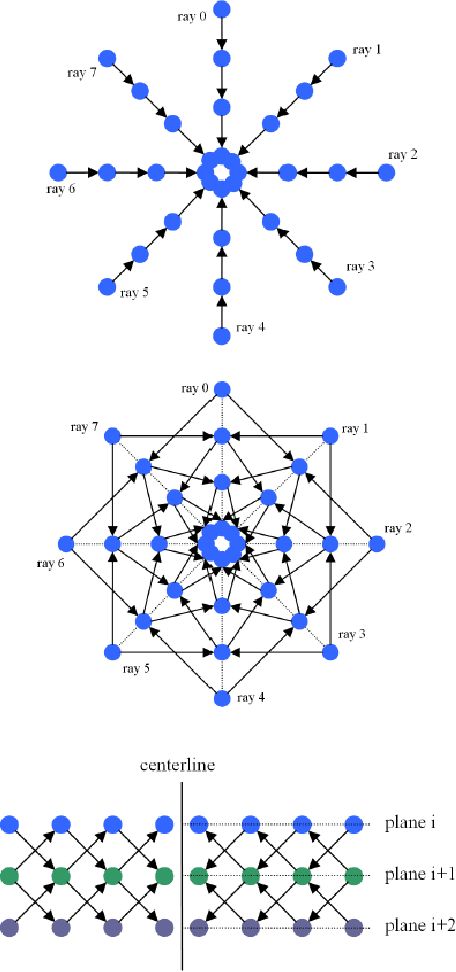
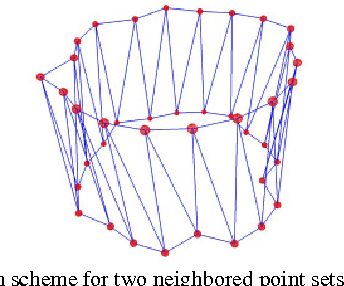
Abstract:Diffusion Tensor Imaging (DTI) provides the possibility of estimating the location and course of eloquent structures in the human brain. Knowledge about this is of high importance for preoperative planning of neurosurgical interventions and for intraoperative guidance by neuronavigation in order to minimize postoperative neurological deficits. Therefore, the segmentation of these structures as closed, three-dimensional object is necessary. In this contribution, two methods for fiber bundle segmentation between two defined regions are compared using software phantoms (abstract model and anatomical phantom modeling the right corticospinal tract). One method uses evaluation points from sampled rays as candidates for boundary points, the other method sets up a directed and weighted (depending on a scalar measure) graph and performs a min-cut for optimal segmentation results. Comparison is done by using the Dice Similarity Coefficient (DSC), a measure for spatial overlap of different segmentation results.
A Semi-Automatic Graph-Based Approach for Determining the Boundary of Eloquent Fiber Bundles in the Human Brain
Mar 08, 2011Abstract:Diffusion Tensor Imaging (DTI) allows estimating the position, orientation and dimension of bundles of nerve pathways. This non-invasive imaging technique takes advantage of the diffusion of water molecules and determines the diffusion coefficients for every voxel of the data set. The identification of the diffusion coefficients and the derivation of information about fiber bundles is of major interest for planning and performing neurosurgical interventions. To minimize the risk of neural deficits during brain surgery as tumor resection (e.g. glioma), the segmentation and integration of the results in the operating room is of prime importance. In this contribution, a robust and efficient graph-based approach for segmentating tubular fiber bundles in the human brain is presented. To define a cost function, the fractional anisotropy (FA) is used, derived from the DTI data, but this value may differ from patient to patient. Besides manually definining seed regions describing the structure of interest, additionally a manual definition of the cost function by the user is necessary. To improve the approach the contribution introduces a solution for automatically determining the cost function by using different 3D masks for each individual data set.
Evaluation of a Novel Approach for Automatic Volume Determination of Glioblastomas Based on Several Manual Expert Segmentations
Mar 08, 2011Abstract:The glioblastoma multiforme is the most common malignant primary brain tumor and is one of the highest malignant human neoplasms. During the course of disease, the evaluation of tumor volume is an essential part of the clinical follow-up. However, manual segmentation for acquisition of tumor volume is a time-consuming process. In this paper, a new approach for the automatic segmentation and volume determination of glioblastomas (glioblastoma multiforme) is presented and evaluated. The approach uses a user-defined seed point inside the glioma to set up a directed 3D graph. The nodes of the graph are obtained by sampling along rays that are sent through the surface points of a polyhedron. After the graph has been constructed, the minimal s-t cut is calculated to separate the glioblastoma from the background. For evaluation, 12 Magnetic Resonance Imaging (MRI) data sets were manually segmented slice by slice, by neurosurgeons with several years of experience in the resection of gliomas. Afterwards, the manual segmentations were compared with the results of the presented approach via the Dice Similarity Coefficient (DSC). For a better assessment of the DSC results, the manual segmentations of the experts were also compared with each other and evaluated via the DSC. In addition, the 12 data sets were segmented once again by one of the neurosurgeons after a period of two weeks, to also measure the intra-physician deviation of the DSC.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge