Dženan Zukić
MedShapeNet -- A Large-Scale Dataset of 3D Medical Shapes for Computer Vision
Sep 12, 2023



Abstract:We present MedShapeNet, a large collection of anatomical shapes (e.g., bones, organs, vessels) and 3D surgical instrument models. Prior to the deep learning era, the broad application of statistical shape models (SSMs) in medical image analysis is evidence that shapes have been commonly used to describe medical data. Nowadays, however, state-of-the-art (SOTA) deep learning algorithms in medical imaging are predominantly voxel-based. In computer vision, on the contrary, shapes (including, voxel occupancy grids, meshes, point clouds and implicit surface models) are preferred data representations in 3D, as seen from the numerous shape-related publications in premier vision conferences, such as the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), as well as the increasing popularity of ShapeNet (about 51,300 models) and Princeton ModelNet (127,915 models) in computer vision research. MedShapeNet is created as an alternative to these commonly used shape benchmarks to facilitate the translation of data-driven vision algorithms to medical applications, and it extends the opportunities to adapt SOTA vision algorithms to solve critical medical problems. Besides, the majority of the medical shapes in MedShapeNet are modeled directly on the imaging data of real patients, and therefore it complements well existing shape benchmarks comprising of computer-aided design (CAD) models. MedShapeNet currently includes more than 100,000 medical shapes, and provides annotations in the form of paired data. It is therefore also a freely available repository of 3D models for extended reality (virtual reality - VR, augmented reality - AR, mixed reality - MR) and medical 3D printing. This white paper describes in detail the motivations behind MedShapeNet, the shape acquisition procedures, the use cases, as well as the usage of the online shape search portal: https://medshapenet.ikim.nrw/
A Comparison of Two Human Brain Tumor Segmentation Methods for MRI Data
Mar 10, 2011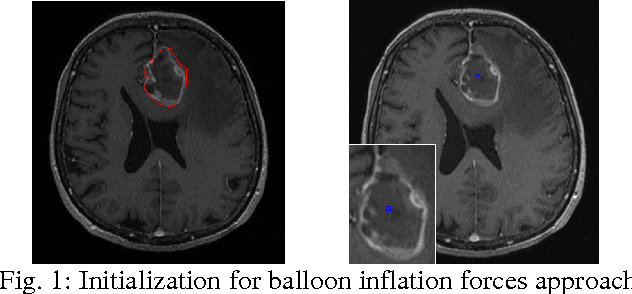
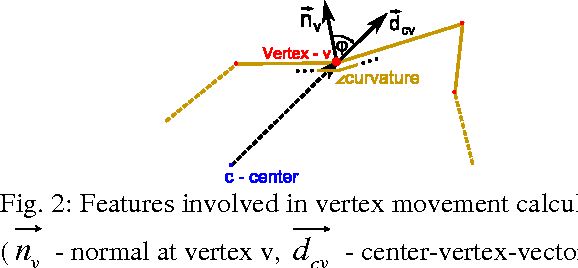
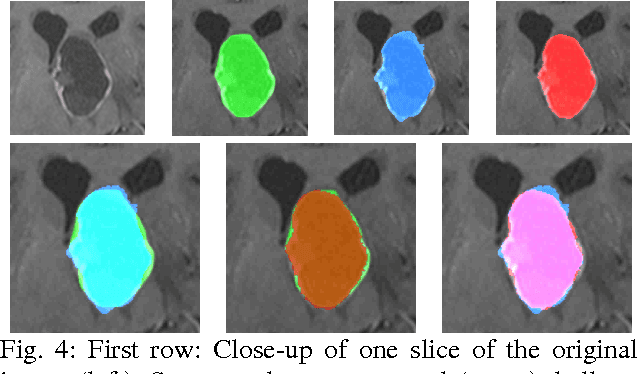
Abstract:The most common primary brain tumors are gliomas, evolving from the cerebral supportive cells. For clinical follow-up, the evaluation of the preoperative tumor volume is essential. Volumetric assessment of tumor volume with manual segmentation of its outlines is a time-consuming process that can be overcome with the help of computerized segmentation methods. In this contribution, two methods for World Health Organization (WHO) grade IV glioma segmentation in the human brain are compared using magnetic resonance imaging (MRI) patient data from the clinical routine. One method uses balloon inflation forces, and relies on detection of high intensity tumor boundaries that are coupled with the use of contrast agent gadolinium. The other method sets up a directed and weighted graph and performs a min-cut for optimal segmentation results. The ground truth of the tumor boundaries - for evaluating the methods on 27 cases - is manually extracted by neurosurgeons with several years of experience in the resection of gliomas. A comparison is performed using the Dice Similarity Coefficient (DSC), a measure for the spatial overlap of different segmentation results.
A Neural Network Classifier of Volume Datasets
Jun 12, 2009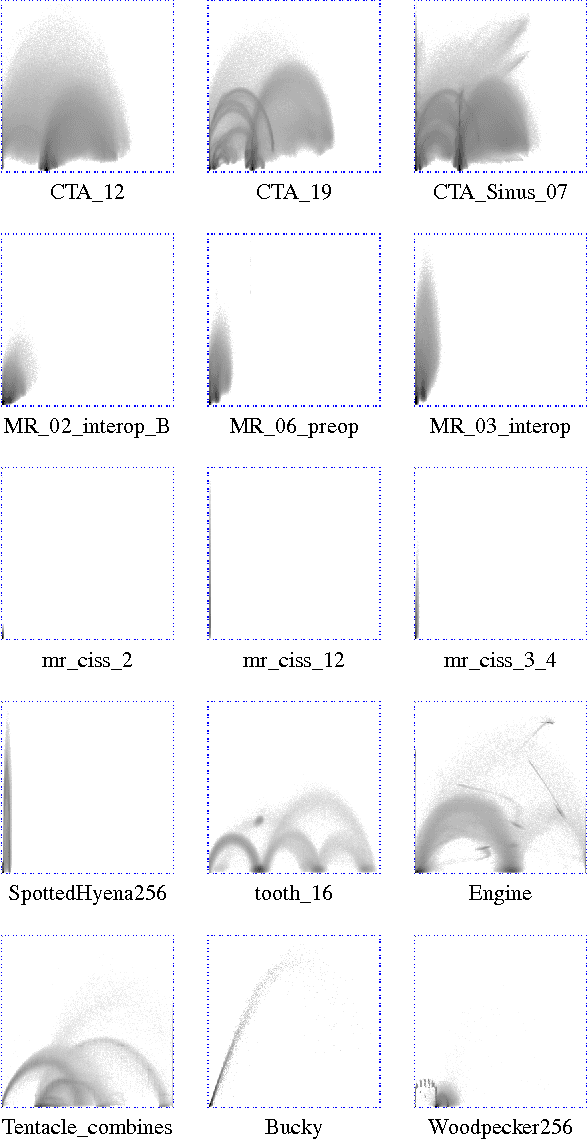
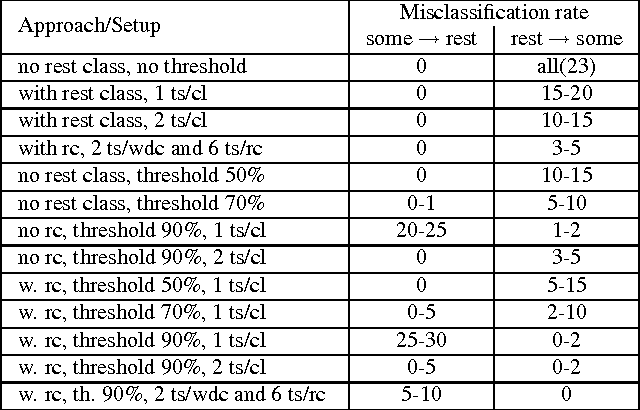

Abstract:Many state-of-the art visualization techniques must be tailored to the specific type of dataset, its modality (CT, MRI, etc.), the recorded object or anatomical region (head, spine, abdomen, etc.) and other parameters related to the data acquisition process. While parts of the information (imaging modality and acquisition sequence) may be obtained from the meta-data stored with the volume scan, there is important information which is not stored explicitly (anatomical region, tracing compound). Also, meta-data might be incomplete, inappropriate or simply missing. This paper presents a novel and simple method of determining the type of dataset from previously defined categories. 2D histograms based on intensity and gradient magnitude of datasets are used as input to a neural network, which classifies it into one of several categories it was trained with. The proposed method is an important building block for visualization systems to be used autonomously by non-experts. The method has been tested on 80 datasets, divided into 3 classes and a "rest" class. A significant result is the ability of the system to classify datasets into a specific class after being trained with only one dataset of that class. Other advantages of the method are its easy implementation and its high computational performance.
* 10 pages, 10 figures, 1 table, 3IA conference http://3ia.teiath.gr/
Neural networks in 3D medical scan visualization
Jun 12, 2009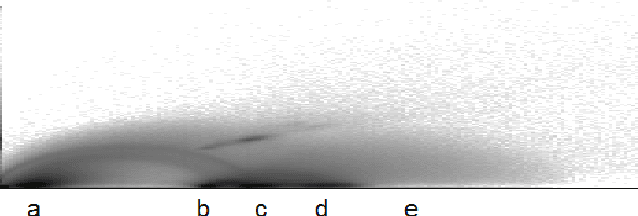

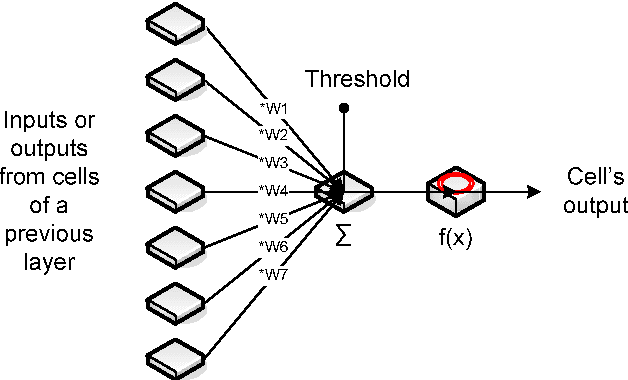
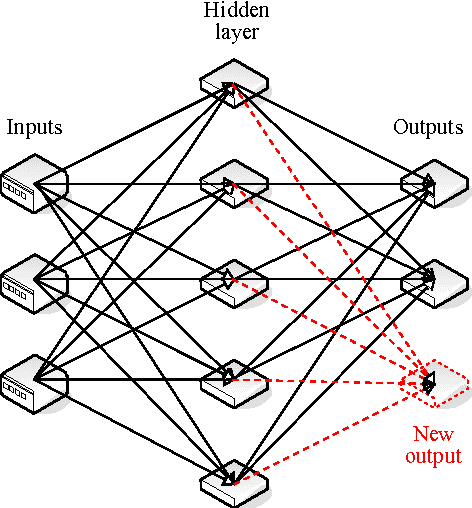
Abstract:For medical volume visualization, one of the most important tasks is to reveal clinically relevant details from the 3D scan (CT, MRI ...), e.g. the coronary arteries, without obscuring them with less significant parts. These volume datasets contain different materials which are difficult to extract and visualize with 1D transfer functions based solely on the attenuation coefficient. Multi-dimensional transfer functions allow a much more precise classification of data which makes it easier to separate different surfaces from each other. Unfortunately, setting up multi-dimensional transfer functions can become a fairly complex task, generally accomplished by trial and error. This paper explains neural networks, and then presents an efficient way to speed up visualization process by semi-automatic transfer function generation. We describe how to use neural networks to detect distinctive features shown in the 2D histogram of the volume data and how to use this information for data classification.
* 8 pages, 6 figures published on conference 3IA'2008 in Athens, Greece (http://3ia.teiath.gr)
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge