Chenglin Wang
ComplexBench-Edit: Benchmarking Complex Instruction-Driven Image Editing via Compositional Dependencies
Jun 15, 2025Abstract:Text-driven image editing has achieved remarkable success in following single instructions. However, real-world scenarios often involve complex, multi-step instructions, particularly ``chain'' instructions where operations are interdependent. Current models struggle with these intricate directives, and existing benchmarks inadequately evaluate such capabilities. Specifically, they often overlook multi-instruction and chain-instruction complexities, and common consistency metrics are flawed. To address this, we introduce ComplexBench-Edit, a novel benchmark designed to systematically assess model performance on complex, multi-instruction, and chain-dependent image editing tasks. ComplexBench-Edit also features a new vision consistency evaluation method that accurately assesses non-modified regions by excluding edited areas. Furthermore, we propose a simple yet powerful Chain-of-Thought (CoT)-based approach that significantly enhances the ability of existing models to follow complex instructions. Our extensive experiments demonstrate ComplexBench-Edit's efficacy in differentiating model capabilities and highlight the superior performance of our CoT-based method in handling complex edits. The data and code are released at https://github.com/llllly26/ComplexBench-Edit.
Diffusion Model with Representation Alignment for Protein Inverse Folding
Dec 12, 2024



Abstract:Protein inverse folding is a fundamental problem in bioinformatics, aiming to recover the amino acid sequences from a given protein backbone structure. Despite the success of existing methods, they struggle to fully capture the intricate inter-residue relationships critical for accurate sequence prediction. We propose a novel method that leverages diffusion models with representation alignment (DMRA), which enhances diffusion-based inverse folding by (1) proposing a shared center that aggregates contextual information from the entire protein structure and selectively distributes it to each residue; and (2) aligning noisy hidden representations with clean semantic representations during the denoising process. This is achieved by predefined semantic representations for amino acid types and a representation alignment method that utilizes type embeddings as semantic feedback to normalize each residue. In experiments, we conduct extensive evaluations on the CATH4.2 dataset to demonstrate that DMRA outperforms leading methods, achieving state-of-the-art performance and exhibiting strong generalization capabilities on the TS50 and TS500 datasets.
A Centralized-Distributed Transfer Model for Cross-Domain Recommendation Based on Multi-Source Heterogeneous Transfer Learning
Nov 14, 2024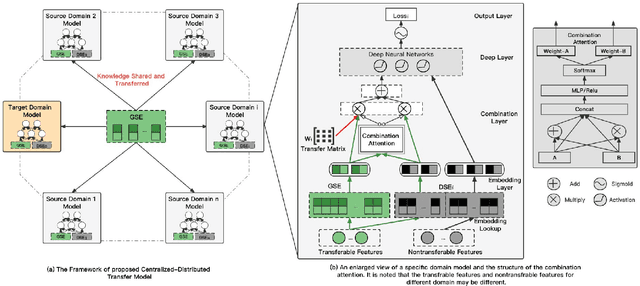
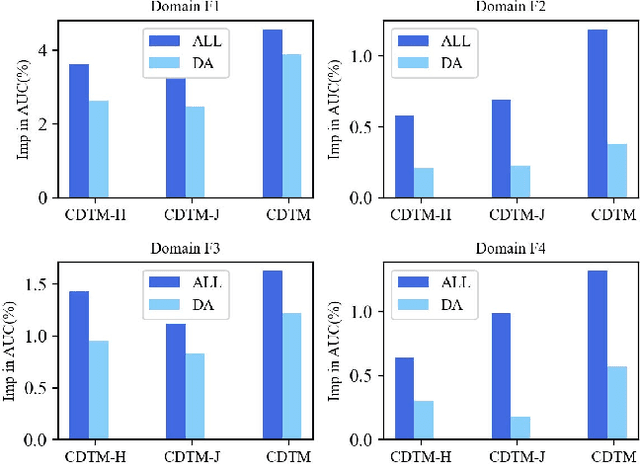
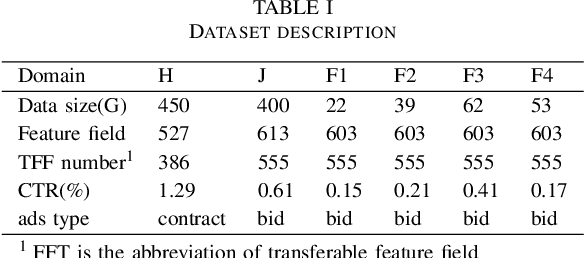
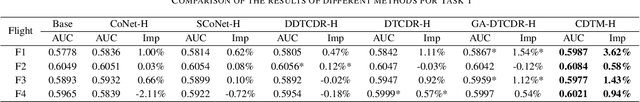
Abstract:Cross-domain recommendation (CDR) methods are proposed to tackle the sparsity problem in click through rate (CTR) estimation. Existing CDR methods directly transfer knowledge from the source domains to the target domain and ignore the heterogeneities among domains, including feature dimensional heterogeneity and latent space heterogeneity, which may lead to negative transfer. Besides, most of the existing methods are based on single-source transfer, which cannot simultaneously utilize knowledge from multiple source domains to further improve the model performance in the target domain. In this paper, we propose a centralized-distributed transfer model (CDTM) for CDR based on multi-source heterogeneous transfer learning. To address the issue of feature dimension heterogeneity, we build a dual embedding structure: domain specific embedding (DSE) and global shared embedding (GSE) to model the feature representation in the single domain and the commonalities in the global space,separately. To solve the latent space heterogeneity, the transfer matrix and attention mechanism are used to map and combine DSE and GSE adaptively. Extensive offline and online experiments demonstrate the effectiveness of our model.
InsectMamba: Insect Pest Classification with State Space Model
Apr 04, 2024



Abstract:The classification of insect pests is a critical task in agricultural technology, vital for ensuring food security and environmental sustainability. However, the complexity of pest identification, due to factors like high camouflage and species diversity, poses significant obstacles. Existing methods struggle with the fine-grained feature extraction needed to distinguish between closely related pest species. Although recent advancements have utilized modified network structures and combined deep learning approaches to improve accuracy, challenges persist due to the similarity between pests and their surroundings. To address this problem, we introduce InsectMamba, a novel approach that integrates State Space Models (SSMs), Convolutional Neural Networks (CNNs), Multi-Head Self-Attention mechanism (MSA), and Multilayer Perceptrons (MLPs) within Mix-SSM blocks. This integration facilitates the extraction of comprehensive visual features by leveraging the strengths of each encoding strategy. A selective module is also proposed to adaptively aggregate these features, enhancing the model's ability to discern pest characteristics. InsectMamba was evaluated against strong competitors across five insect pest classification datasets. The results demonstrate its superior performance and verify the significance of each model component by an ablation study.
Unsupervised Knowledge Graph Construction and Event-centric Knowledge Infusion for Scientific NLI
Oct 28, 2022



Abstract:With the advance of natural language inference (NLI), a rising demand for NLI is to handle scientific texts. Existing methods depend on pre-trained models (PTM) which lack domain-specific knowledge. To tackle this drawback, we introduce a scientific knowledge graph to generalize PTM to scientific domain. However, existing knowledge graph construction approaches suffer from some drawbacks, i.e., expensive labeled data, failure to apply in other domains, long inference time and difficulty extending to large corpora. Therefore, we propose an unsupervised knowledge graph construction method to build a scientific knowledge graph (SKG) without any labeled data. Moreover, to alleviate noise effect from SKG and complement knowledge in sentences better, we propose an event-centric knowledge infusion method to integrate external knowledge into each event that is a fine-grained semantic unit in sentences. Experimental results show that our method achieves state-of-the-art performance and the effectiveness and reliability of SKG.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge