Chaotao Ding
Syllables to Scenes: Literary-Guided Free-Viewpoint 3D Scene Synthesis from Japanese Haiku
Feb 17, 2025Abstract:In the era of the metaverse, where immersive technologies redefine human experiences, translating abstract literary concepts into navigable 3D environments presents a fundamental challenge in preserving semantic and emotional fidelity. This research introduces HaikuVerse, a novel framework for transforming poetic abstraction into spatial representation, with Japanese Haiku serving as an ideal test case due to its sophisticated encapsulation of profound emotions and imagery within minimal text. While existing text-to-3D methods struggle with nuanced interpretations, we present a literary-guided approach that synergizes traditional poetry analysis with advanced generative technologies. Our framework centers on two key innovations: (1) Hierarchical Literary-Criticism Theory Grounded Parsing (H-LCTGP), which captures both explicit imagery and implicit emotional resonance through structured semantic decomposition, and (2) Progressive Dimensional Synthesis (PDS), a multi-stage pipeline that systematically transforms poetic elements into coherent 3D scenes through sequential diffusion processes, geometric optimization, and real-time enhancement. Extensive experiments demonstrate that HaikuVerse significantly outperforms conventional text-to-3D approaches in both literary fidelity and visual quality, establishing a new paradigm for preserving cultural heritage in immersive digital spaces. Project website at: https://syllables-to-scenes.github.io/
SAM2-Adapter: Evaluating & Adapting Segment Anything 2 in Downstream Tasks: Camouflage, Shadow, Medical Image Segmentation, and More
Aug 08, 2024Abstract:The advent of large models, also known as foundation models, has significantly transformed the AI research landscape, with models like Segment Anything (SAM) achieving notable success in diverse image segmentation scenarios. Despite its advancements, SAM encountered limitations in handling some complex low-level segmentation tasks like camouflaged object and medical imaging. In response, in 2023, we introduced SAM-Adapter, which demonstrated improved performance on these challenging tasks. Now, with the release of Segment Anything 2 (SAM2), a successor with enhanced architecture and a larger training corpus, we reassess these challenges. This paper introduces SAM2-Adapter, the first adapter designed to overcome the persistent limitations observed in SAM2 and achieve new state-of-the-art (SOTA) results in specific downstream tasks including medical image segmentation, camouflaged (concealed) object detection, and shadow detection. SAM2-Adapter builds on the SAM-Adapter's strengths, offering enhanced generalizability and composability for diverse applications. We present extensive experimental results demonstrating SAM2-Adapter's effectiveness. We show the potential and encourage the research community to leverage the SAM2 model with our SAM2-Adapter for achieving superior segmentation outcomes. Code, pre-trained models, and data processing protocols are available at http://tianrun-chen.github.io/SAM-Adaptor/
Magic3DSketch: Create Colorful 3D Models From Sketch-Based 3D Modeling Guided by Text and Language-Image Pre-Training
Jul 27, 2024


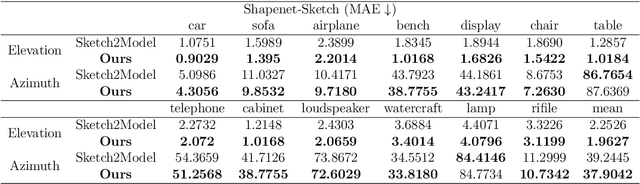
Abstract:The requirement for 3D content is growing as AR/VR application emerges. At the same time, 3D modelling is only available for skillful experts, because traditional methods like Computer-Aided Design (CAD) are often too labor-intensive and skill-demanding, making it challenging for novice users. Our proposed method, Magic3DSketch, employs a novel technique that encodes sketches to predict a 3D mesh, guided by text descriptions and leveraging external prior knowledge obtained through text and language-image pre-training. The integration of language-image pre-trained neural networks complements the sparse and ambiguous nature of single-view sketch inputs. Our method is also more useful and offers higher degree of controllability compared to existing text-to-3D approaches, according to our user study. Moreover, Magic3DSketch achieves state-of-the-art performance in both synthetic and real dataset with the capability of producing more detailed structures and realistic shapes with the help of text input. Users are also more satisfied with models obtained by Magic3DSketch according to our user study. Additionally, we are also the first, to our knowledge, add color based on text description to the sketch-derived shapes. By combining sketches and text guidance with the help of language-image pretrained models, our Magic3DSketch can allow novice users to create custom 3D models with minimal effort and maximum creative freedom, with the potential to revolutionize future 3D modeling pipelines.
xLSTM-UNet can be an Effective 2D & 3D Medical Image Segmentation Backbone with Vision-LSTM (ViL) better than its Mamba Counterpart
Jul 02, 2024Abstract:Convolutional Neural Networks (CNNs) and Vision Transformers (ViT) have been pivotal in biomedical image segmentation, yet their ability to manage long-range dependencies remains constrained by inherent locality and computational overhead. To overcome these challenges, in this technical report, we first propose xLSTM-UNet, a UNet structured deep learning neural network that leverages Vision-LSTM (xLSTM) as its backbone for medical image segmentation. xLSTM is a recently proposed as the successor of Long Short-Term Memory (LSTM) networks and have demonstrated superior performance compared to Transformers and State Space Models (SSMs) like Mamba in Neural Language Processing (NLP) and image classification (as demonstrated in Vision-LSTM, or ViL implementation). Here, xLSTM-UNet we designed extend the success in biomedical image segmentation domain. By integrating the local feature extraction strengths of convolutional layers with the long-range dependency capturing abilities of xLSTM, xLSTM-UNet offers a robust solution for comprehensive image analysis. We validate the efficacy of xLSTM-UNet through experiments. Our findings demonstrate that xLSTM-UNet consistently surpasses the performance of leading CNN-based, Transformer-based, and Mamba-based segmentation networks in multiple datasets in biomedical segmentation including organs in abdomen MRI, instruments in endoscopic images, and cells in microscopic images. With comprehensive experiments performed, this technical report highlights the potential of xLSTM-based architectures in advancing biomedical image analysis in both 2D and 3D. The code, models, and datasets are publicly available at http://tianrun-chen.github.io/xLSTM-UNet/
SAM Fails to Segment Anything? -- SAM-Adapter: Adapting SAM in Underperformed Scenes: Camouflage, Shadow, Medical Image Segmentation, and More
May 02, 2023Abstract:The emergence of large models, also known as foundation models, has brought significant advancements to AI research. One such model is Segment Anything (SAM), which is designed for image segmentation tasks. However, as with other foundation models, our experimental findings suggest that SAM may fail or perform poorly in certain segmentation tasks, such as shadow detection and camouflaged object detection (concealed object detection). This study first paves the way for applying the large pre-trained image segmentation model SAM to these downstream tasks, even in situations where SAM performs poorly. Rather than fine-tuning the SAM network, we propose \textbf{SAM-Adapter}, which incorporates domain-specific information or visual prompts into the segmentation network by using simple yet effective adapters. By integrating task-specific knowledge with general knowledge learnt by the large model, SAM-Adapter can significantly elevate the performance of SAM in challenging tasks as shown in extensive experiments. We can even outperform task-specific network models and achieve state-of-the-art performance in the task we tested: camouflaged object detection, shadow detection. We also tested polyp segmentation (medical image segmentation) and achieves better results. We believe our work opens up opportunities for utilizing SAM in downstream tasks, with potential applications in various fields, including medical image processing, agriculture, remote sensing, and more.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge