Caroline Chung
MLPrE -- A tool for preprocessing and exploratory data analysis prior to machine learning model construction
Oct 29, 2025Abstract:With the recent growth of Deep Learning for AI, there is a need for tools to meet the demand of data flowing into those models. In some cases, source data may exist in multiple formats, and therefore the source data must be investigated and properly engineered for a Machine Learning model or graph database. Overhead and lack of scalability with existing workflows limit integration within a larger processing pipeline such as Apache Airflow, driving the need for a robust, extensible, and lightweight tool to preprocess arbitrary datasets that scales with data type and size. To address this, we present Machine Learning Preprocessing and Exploratory Data Analysis, MLPrE, in which SparkDataFrames were utilized to hold data during processing and ensure scalability. A generalizable JSON input file format was utilized to describe stepwise changes to that DataFrame. Stages were implemented for input and output, filtering, basic statistics, feature engineering, and exploratory data analysis. A total of 69 stages were implemented into MLPrE, of which we highlight and demonstrate key stages using six diverse datasets. We further highlight MLPrE's ability to independently process multiple fields in flat files and recombine them, otherwise requiring an additional pipeline, using a UniProt glossary term dataset. Building on this advantage, we demonstrated the clustering stage with available wine quality data. Lastly, we demonstrate the preparation of data for a graph database in the final stages of MLPrE using phosphosite kinase data. Overall, our MLPrE tool offers a generalizable and scalable tool for preprocessing and early data analysis, filling a critical need for such a tool given the ever expanding use of machine learning. This tool serves to accelerate and simplify early stage development in larger workflows.
MIST: A Simple and Scalable End-To-End 3D Medical Imaging Segmentation Framework
Jul 31, 2024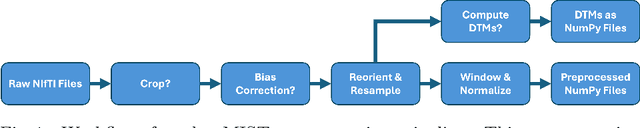
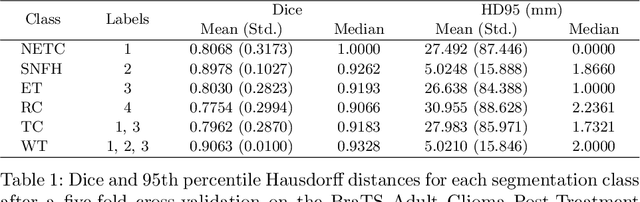
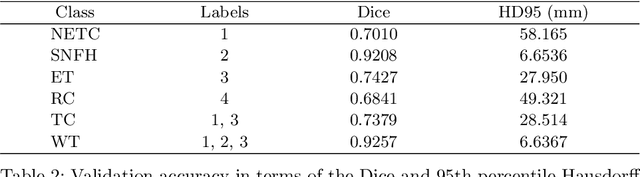
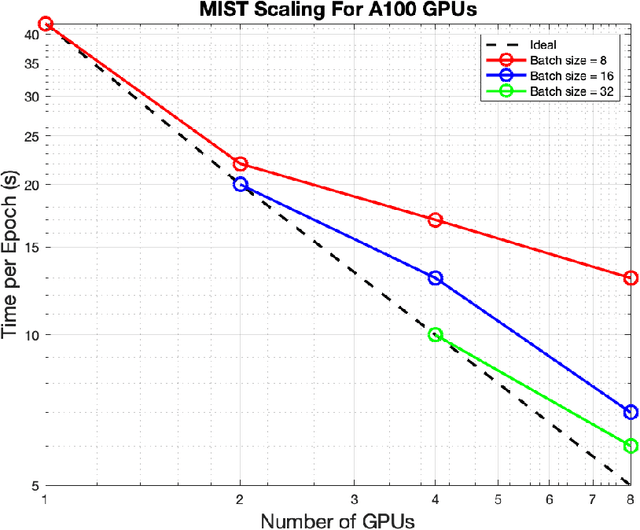
Abstract:Medical imaging segmentation is a highly active area of research, with deep learning-based methods achieving state-of-the-art results in several benchmarks. However, the lack of standardized tools for training, testing, and evaluating new methods makes the comparison of methods difficult. To address this, we introduce the Medical Imaging Segmentation Toolkit (MIST), a simple, modular, and end-to-end medical imaging segmentation framework designed to facilitate consistent training, testing, and evaluation of deep learning-based medical imaging segmentation methods. MIST standardizes data analysis, preprocessing, and evaluation pipelines, accommodating multiple architectures and loss functions. This standardization ensures reproducible and fair comparisons across different methods. We detail MIST's data format requirements, pipelines, and auxiliary features and demonstrate its efficacy using the BraTS Adult Glioma Post-Treatment Challenge dataset. Our results highlight MIST's ability to produce accurate segmentation masks and its scalability across multiple GPUs, showcasing its potential as a powerful tool for future medical imaging research and development.
Evaluating the Performance of StyleGAN2-ADA on Medical Images
Oct 07, 2022Abstract:Although generative adversarial networks (GANs) have shown promise in medical imaging, they have four main limitations that impeded their utility: computational cost, data requirements, reliable evaluation measures, and training complexity. Our work investigates each of these obstacles in a novel application of StyleGAN2-ADA to high-resolution medical imaging datasets. Our dataset is comprised of liver-containing axial slices from non-contrast and contrast-enhanced computed tomography (CT) scans. Additionally, we utilized four public datasets composed of various imaging modalities. We trained a StyleGAN2 network with transfer learning (from the Flickr-Faces-HQ dataset) and data augmentation (horizontal flipping and adaptive discriminator augmentation). The network's generative quality was measured quantitatively with the Fr\'echet Inception Distance (FID) and qualitatively with a visual Turing test given to seven radiologists and radiation oncologists. The StyleGAN2-ADA network achieved a FID of 5.22 ($\pm$ 0.17) on our liver CT dataset. It also set new record FIDs of 10.78, 3.52, 21.17, and 5.39 on the publicly available SLIVER07, ChestX-ray14, ACDC, and Medical Segmentation Decathlon (brain tumors) datasets. In the visual Turing test, the clinicians rated generated images as real 42% of the time, approaching random guessing. Our computational ablation study revealed that transfer learning and data augmentation stabilize training and improve the perceptual quality of the generated images. We observed the FID to be consistent with human perceptual evaluation of medical images. Finally, our work found that StyleGAN2-ADA consistently produces high-quality results without hyperparameter searches or retraining.
* This preprint has not undergone post-submission improvements or corrections. The Version of Record of this contribution is published in LNCS, volume 13570, and is available online at https://doi.org/10.1007/978-3-031-16980-9_14
Integrative Imaging Informatics for Cancer Research: Workflow Automation for Neuro-oncology (I3CR-WANO)
Oct 06, 2022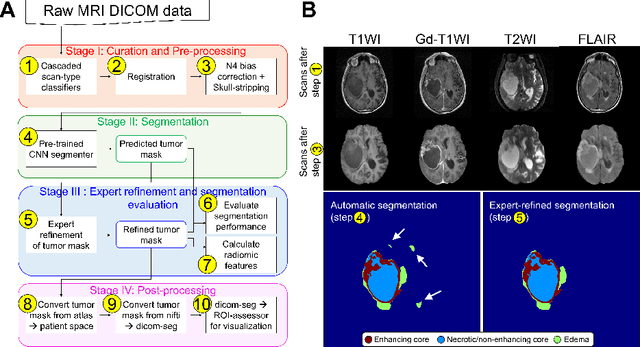
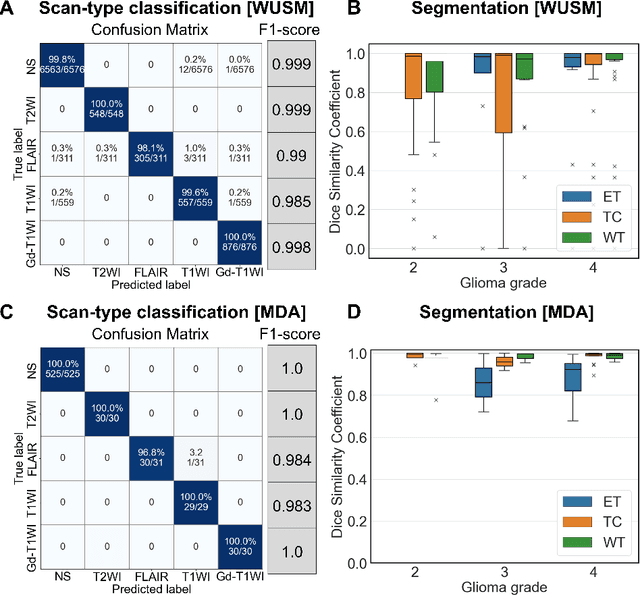
Abstract:Efforts to utilize growing volumes of clinical imaging data to generate tumor evaluations continue to require significant manual data wrangling owing to the data heterogeneity. Here, we propose an artificial intelligence-based solution for the aggregation and processing of multisequence neuro-oncology MRI data to extract quantitative tumor measurements. Our end-to-end framework i) classifies MRI sequences using an ensemble classifier, ii) preprocesses the data in a reproducible manner, iii) delineates tumor tissue subtypes using convolutional neural networks, and iv) extracts diverse radiomic features. Moreover, it is robust to missing sequences and adopts an expert-in-the-loop approach, where the segmentation results may be manually refined by radiologists. Following the implementation of the framework in Docker containers, it was applied to two retrospective glioma datasets collected from the Washington University School of Medicine (WUSM; n = 384) and the M.D. Anderson Cancer Center (MDA; n = 30) comprising preoperative MRI scans from patients with pathologically confirmed gliomas. The scan-type classifier yielded an accuracy of over 99%, correctly identifying sequences from 380/384 and 30/30 sessions from the WUSM and MDA datasets, respectively. Segmentation performance was quantified using the Dice Similarity Coefficient between the predicted and expert-refined tumor masks. Mean Dice scores were 0.882 ($\pm$0.244) and 0.977 ($\pm$0.04) for whole tumor segmentation for WUSM and MDA, respectively. This streamlined framework automatically curated, processed, and segmented raw MRI data of patients with varying grades of gliomas, enabling the curation of large-scale neuro-oncology datasets and demonstrating a high potential for integration as an assistive tool in clinical practice.
Automated WBRT Treatment Planning via Deep Learning Auto-Contouring and Customizable Landmark-Based Field Aperture Design
May 24, 2022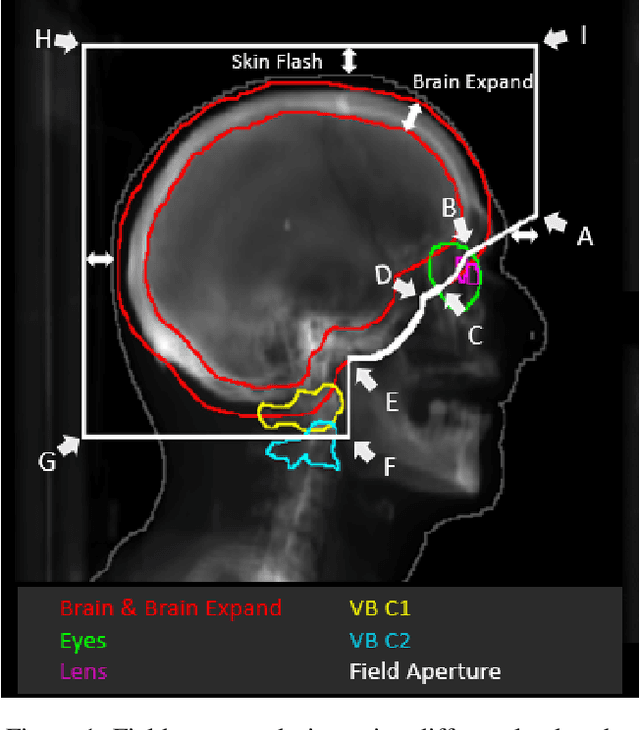


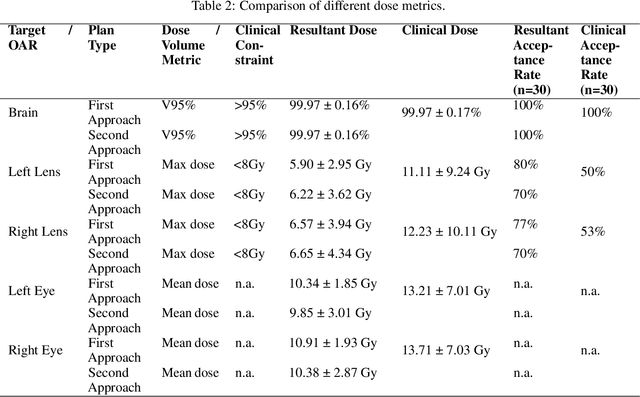
Abstract:In this work, we developed and evaluated a novel pipeline consisting of two landmark-based field aperture generation approaches for WBRT treatment planning; they are fully automated and customizable. The automation pipeline is beneficial for both clinicians and patients, where we can reduce clinician workload and reduce treatment planning time. The customizability of the field aperture design addresses different clinical requirements and allows the personalized design to become feasible. The performance results regarding quantitative and qualitative evaluations demonstrated that our plans were comparable with the original clinical plans. This technique has been deployed as part of a fully automated treatment planning tool for whole-brain cancer and could be translated to other treatment sites in the future.
Correlation between image quality metrics of magnetic resonance images and the neural network segmentation accuracy
Nov 01, 2021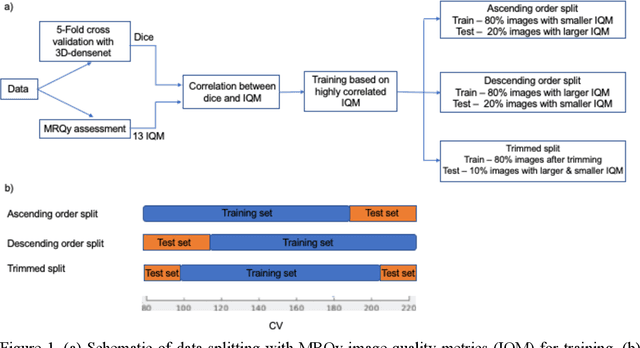
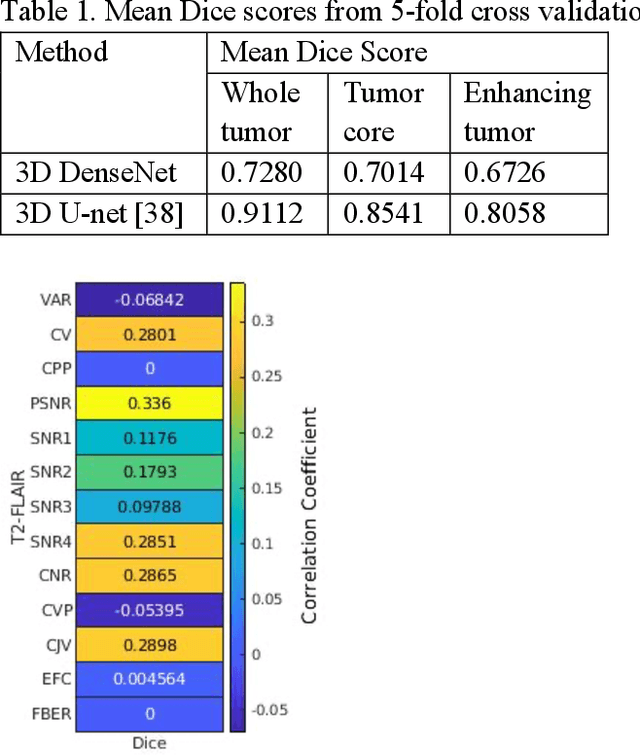
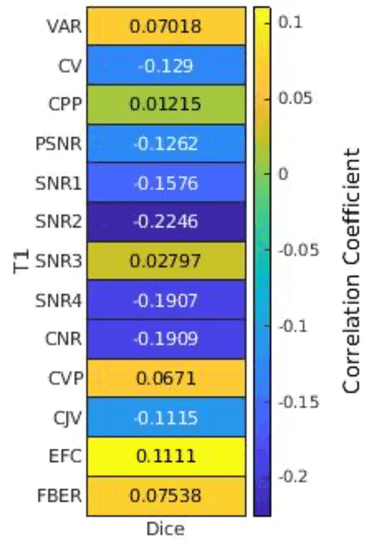
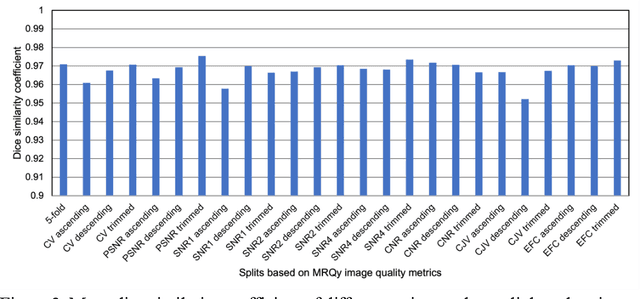
Abstract:Deep neural networks with multilevel connections process input data in complex ways to learn the information.A networks learning efficiency depends not only on the complex neural network architecture but also on the input training images.Medical image segmentation with deep neural networks for skull stripping or tumor segmentation from magnetic resonance images enables learning both global and local features of the images.Though medical images are collected in a controlled environment,there may be artifacts or equipment based variance that cause inherent bias in the input set.In this study, we investigated the correlation between the image quality metrics of MR images with the neural network segmentation accuracy.For that we have used the 3D DenseNet architecture and let the network trained on the same input but applying different methodologies to select the training data set based on the IQM values.The difference in the segmentation accuracy between models based on the random training inputs with IQM based training inputs shed light on the role of image quality metrics on segmentation accuracy.By running the image quality metrics to choose the training inputs,further we may tune the learning efficiency of the network and the segmentation accuracy.
PocketNet: A Smaller Neural Network for 3D Medical Image Segmentation
Apr 21, 2021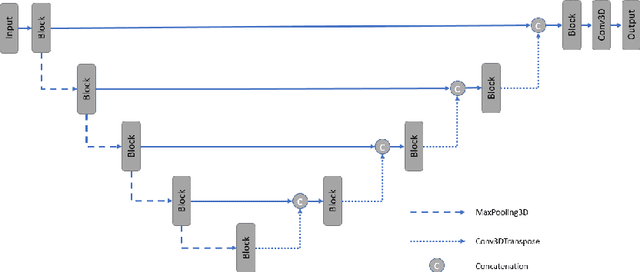
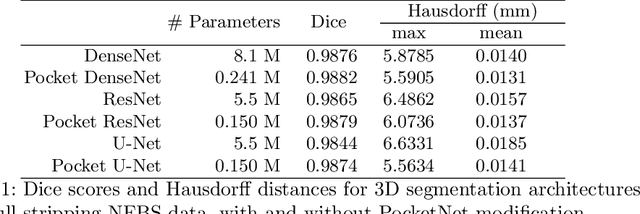
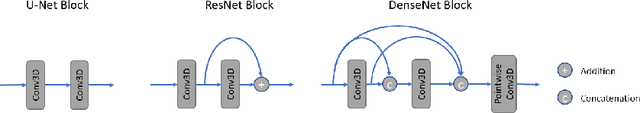
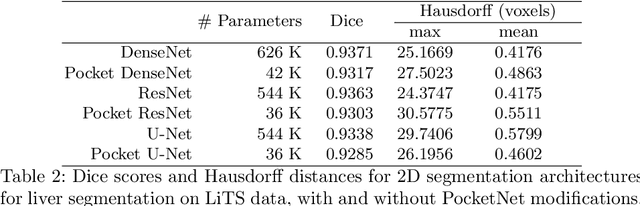
Abstract:Overparameterized deep learning networks have shown impressive performance in the area of automatic medical image segmentation. However, they achieve this performance at an enormous cost in memory, runtime, and energy. A large source of overparameterization in modern neural networks results from doubling the number of feature maps with each downsampling layer. This rapid growth in the number of parameters results in network architectures that require a significant amount of computing resources, making them less accessible and difficult to use. By keeping the number of feature maps constant throughout the network, we derive a new CNN architecture called PocketNet that achieves comparable segmentation results to conventional CNNs while using less than 3% of the number of parameters.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge