David Fuentes
A Priori Generalizability Estimate for a CNN
Feb 24, 2025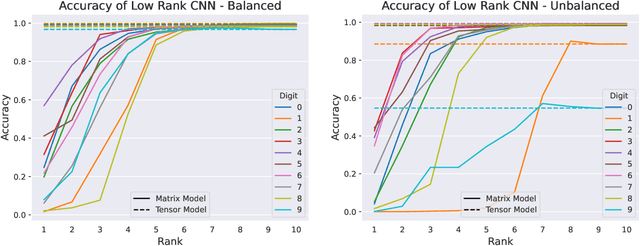
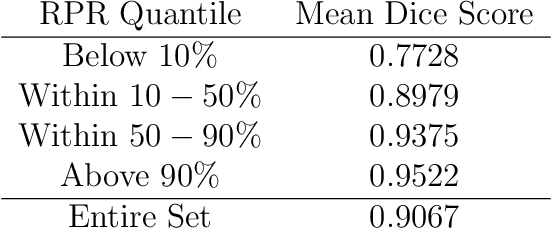
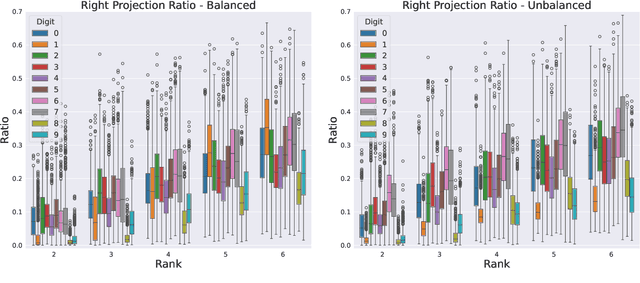
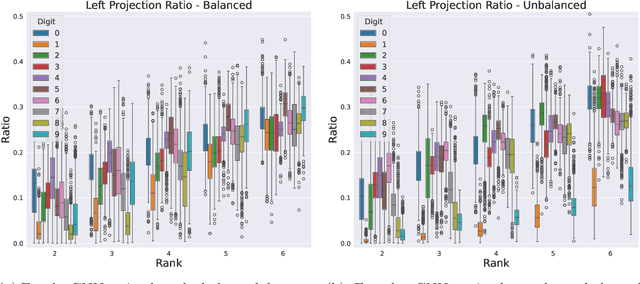
Abstract:We formulate truncated singular value decompositions of entire convolutional neural networks. We demonstrate the computed left and right singular vectors are useful in identifying which images the convolutional neural network is likely to perform poorly on. To create this diagnostic tool, we define two metrics: the Right Projection Ratio and the Left Projection Ratio. The Right (Left) Projection Ratio evaluates the fidelity of the projection of an image (label) onto the computed right (left) singular vectors. We observe that both ratios are able to identify the presence of class imbalance for an image classification problem. Additionally, the Right Projection Ratio, which only requires unlabeled data, is found to be correlated to the model's performance when applied to image segmentation. This suggests the Right Projection Ratio could be a useful metric to estimate how likely the model is to perform well on a sample.
Learning Discontinuous Galerkin Solutions to Elliptic Problems via Small Linear Convolutional Neural Networks
Feb 12, 2025



Abstract:In recent years, there has been an increasing interest in using deep learning and neural networks to tackle scientific problems, particularly in solving partial differential equations (PDEs). However, many neural network-based methods, such as physics-informed neural networks, depend on automatic differentiation and the sampling of collocation points, which can result in a lack of interpretability and lower accuracy compared to traditional numerical methods. To address this issue, we propose two approaches for learning discontinuous Galerkin solutions to PDEs using small linear convolutional neural networks. Our first approach is supervised and depends on labeled data, while our second approach is unsupervised and does not rely on any training data. In both cases, our methods use substantially fewer parameters than similar numerics-based neural networks while also demonstrating comparable accuracy to the true and DG solutions for elliptic problems.
An Adaptive Collocation Point Strategy For Physics Informed Neural Networks via the QR Discrete Empirical Interpolation Method
Jan 16, 2025



Abstract:Physics-informed neural networks (PINNs) have gained significant attention for solving forward and inverse problems related to partial differential equations (PDEs). While advancements in loss functions and network architectures have improved PINN accuracy, the impact of collocation point sampling on their performance remains underexplored. Fixed sampling methods, such as uniform random sampling and equispaced grids, can fail to capture critical regions with high solution gradients, limiting their effectiveness for complex PDEs. Adaptive methods, inspired by adaptive mesh refinement from traditional numerical methods, address this by dynamically updating collocation points during training but may overlook residual dynamics between updates, potentially losing valuable information. To overcome this limitation, we propose an adaptive collocation point selection strategy utilizing the QR Discrete Empirical Interpolation Method (QR-DEIM), a reduced-order modeling technique for efficiently approximating nonlinear functions. Our results on benchmark PDEs, including the wave, Allen-Cahn, and Burgers' equations, demonstrate that our QR-DEIM-based approach improves PINN accuracy compared to existing methods, offering a promising direction for adaptive collocation point strategies.
Two Stage Segmentation of Cervical Tumors using PocketNet
Sep 17, 2024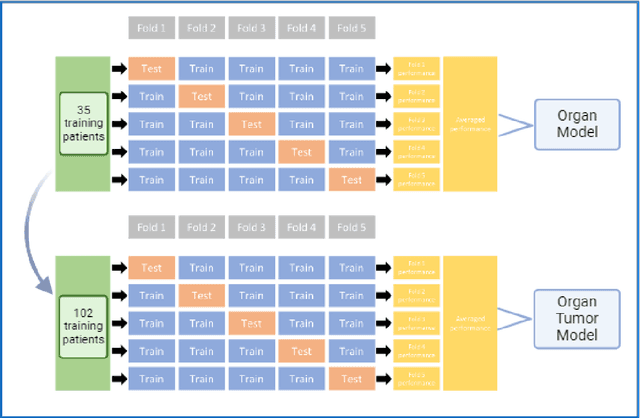

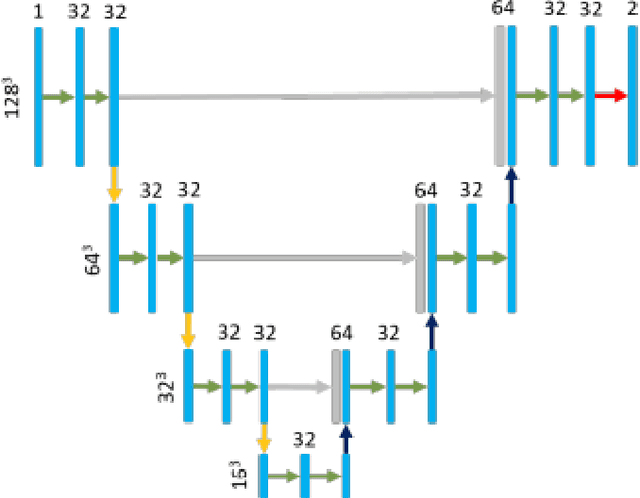
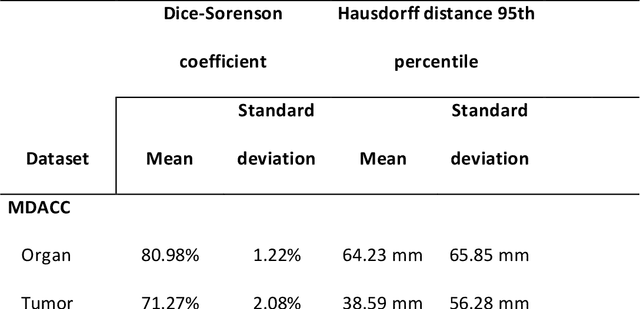
Abstract:Cervical cancer remains the fourth most common malignancy amongst women worldwide.1 Concurrent chemoradiotherapy (CRT) serves as the mainstay definitive treatment regimen for locally advanced cervical cancers and includes external beam radiation followed by brachytherapy.2 Integral to radiotherapy treatment planning is the routine contouring of both the target tumor at the level of the cervix, associated gynecologic anatomy and the adjacent organs at risk (OARs). However, manual contouring of these structures is both time and labor intensive and associated with known interobserver variability that can impact treatment outcomes. While multiple tools have been developed to automatically segment OARs and the high-risk clinical tumor volume (HR-CTV) using computed tomography (CT) images,3,4,5,6 the development of deep learning-based tumor segmentation tools using routine T2-weighted (T2w) magnetic resonance imaging (MRI) addresses an unmet clinical need to improve the routine contouring of both anatomical structures and cervical cancers, thereby increasing quality and consistency of radiotherapy planning. This work applied a novel deep-learning model (PocketNet) to segment the cervix, vagina, uterus, and tumor(s) on T2w MRI. The performance of the PocketNet architecture was evaluated, when trained on data via 5-fold cross validation. PocketNet achieved a mean Dice-Sorensen similarity coefficient (DSC) exceeding 70% for tumor segmentation and 80% for organ segmentation. These results suggest that PocketNet is robust to variations in contrast protocols, providing reliable segmentation of the ROIs.
MIST: A Simple and Scalable End-To-End 3D Medical Imaging Segmentation Framework
Jul 31, 2024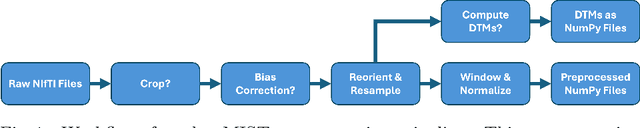
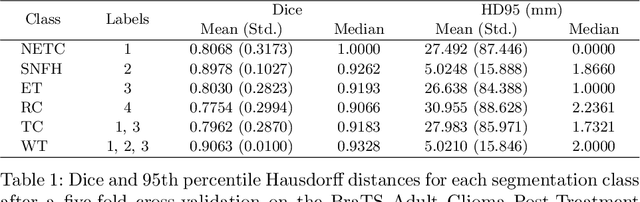
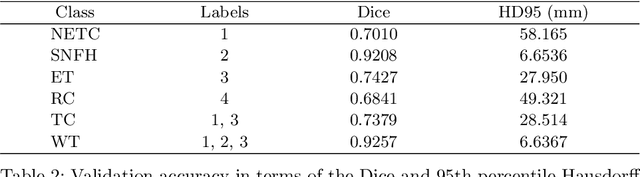
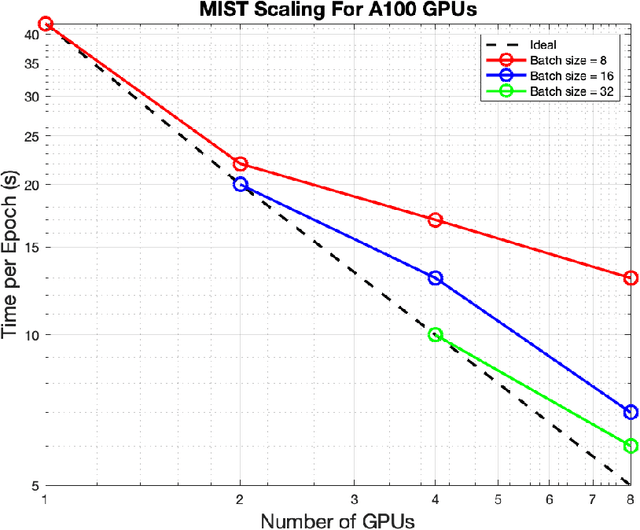
Abstract:Medical imaging segmentation is a highly active area of research, with deep learning-based methods achieving state-of-the-art results in several benchmarks. However, the lack of standardized tools for training, testing, and evaluating new methods makes the comparison of methods difficult. To address this, we introduce the Medical Imaging Segmentation Toolkit (MIST), a simple, modular, and end-to-end medical imaging segmentation framework designed to facilitate consistent training, testing, and evaluation of deep learning-based medical imaging segmentation methods. MIST standardizes data analysis, preprocessing, and evaluation pipelines, accommodating multiple architectures and loss functions. This standardization ensures reproducible and fair comparisons across different methods. We detail MIST's data format requirements, pipelines, and auxiliary features and demonstrate its efficacy using the BraTS Adult Glioma Post-Treatment Challenge dataset. Our results highlight MIST's ability to produce accurate segmentation masks and its scalability across multiple GPUs, showcasing its potential as a powerful tool for future medical imaging research and development.
Solutions to Elliptic and Parabolic Problems via Finite Difference Based Unsupervised Small Linear Convolutional Neural Networks
Nov 01, 2023



Abstract:In recent years, there has been a growing interest in leveraging deep learning and neural networks to address scientific problems, particularly in solving partial differential equations (PDEs). However, current neural network-based PDE solvers often rely on extensive training data or labeled input-output pairs, making them prone to challenges in generalizing to out-of-distribution examples. To mitigate the generalization gap encountered by conventional neural network-based methods in estimating PDE solutions, we formulate a fully unsupervised approach, requiring no training data, to estimate finite difference solutions for PDEs directly via small convolutional neural networks. Our proposed algorithms demonstrate a comparable accuracy to the true solution for several selected elliptic and parabolic problems compared to the finite difference method.
Distributed Conditional GAN (discGAN) For Synthetic Healthcare Data Generation
Apr 09, 2023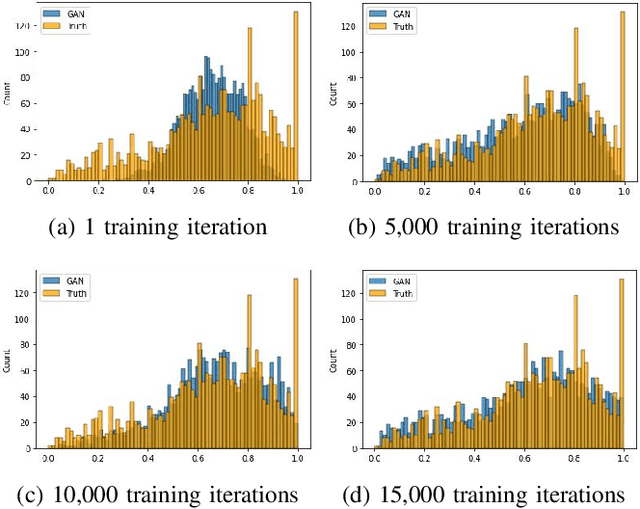
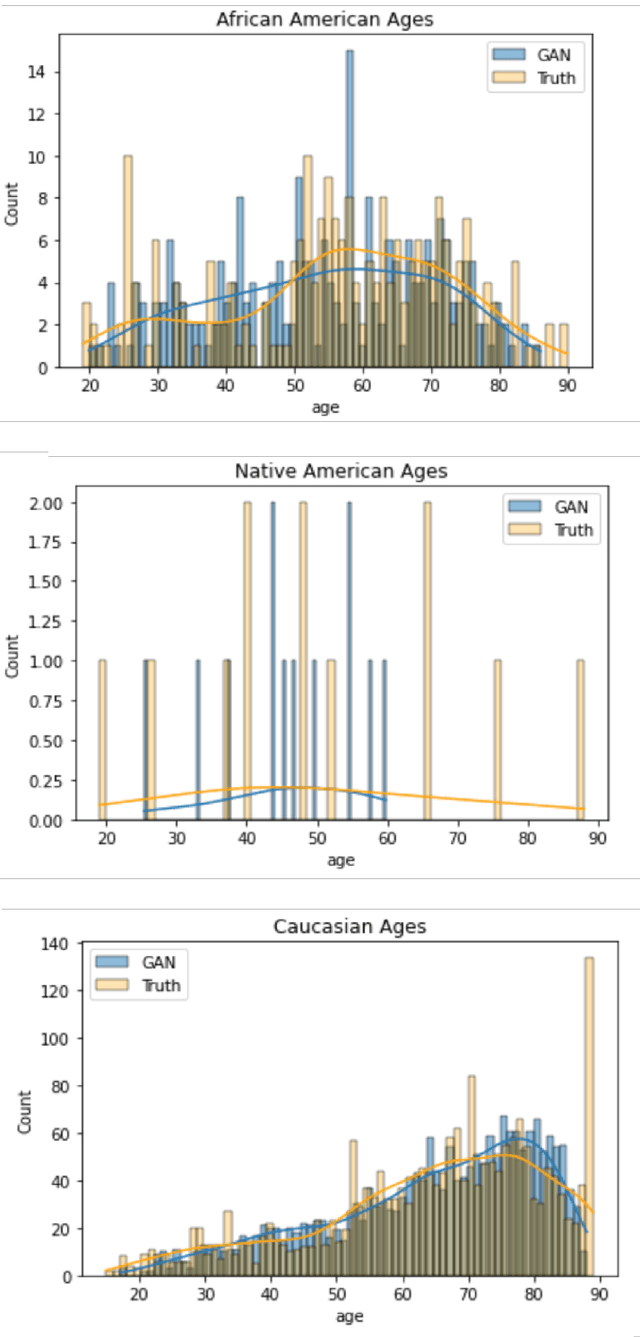
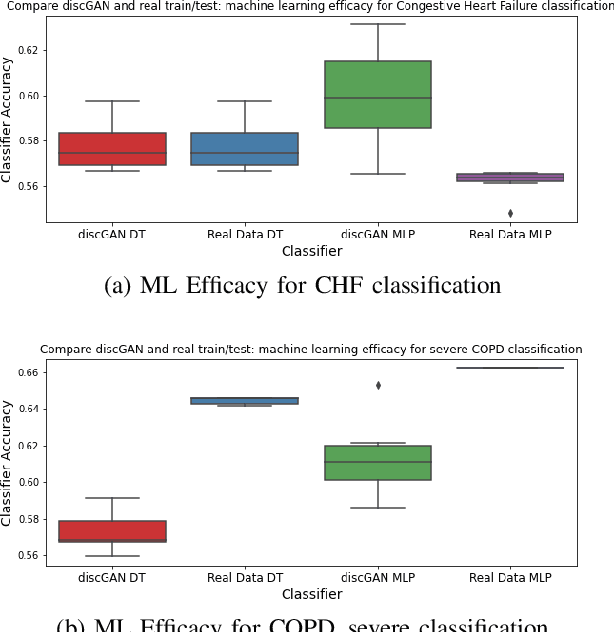
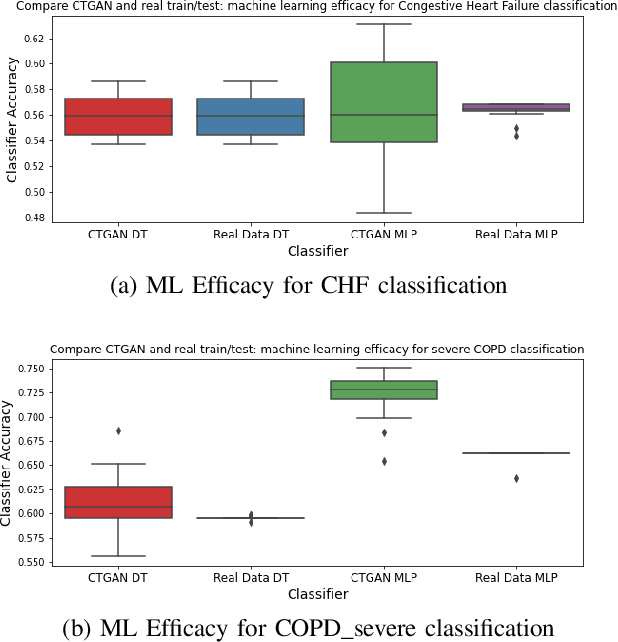
Abstract:In this paper, we propose a distributed Generative Adversarial Networks (discGANs) to generate synthetic tabular data specific to the healthcare domain. While using GANs to generate images has been well studied, little to no attention has been given to generation of tabular data. Modeling distributions of discrete and continuous tabular data is a non-trivial task with high utility. We applied discGAN to model non-Gaussian multi-modal healthcare data. We generated 249,000 synthetic records from original 2,027 eICU dataset. We evaluated the performance of the model using machine learning efficacy, the Kolmogorov-Smirnov (KS) test for continuous variables and chi-squared test for discrete variables. Our results show that discGAN was able to generate data with distributions similar to the real data.
FMG-Net and W-Net: Multigrid Inspired Deep Learning Architectures For Medical Imaging Segmentation
Apr 05, 2023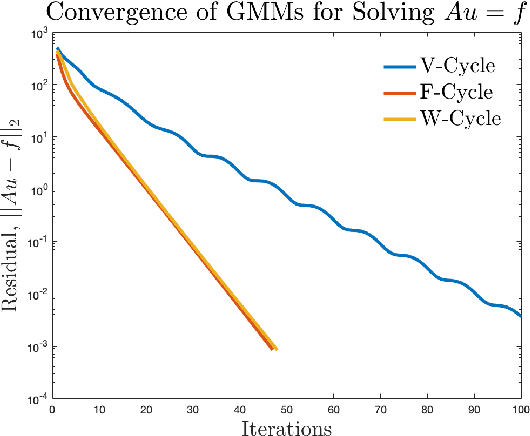
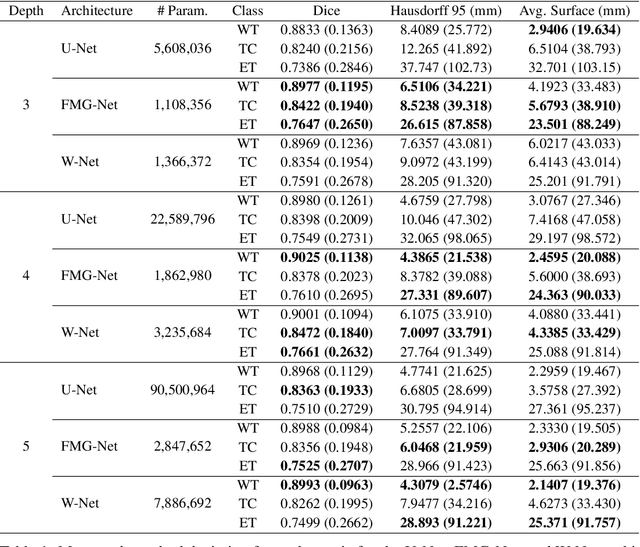
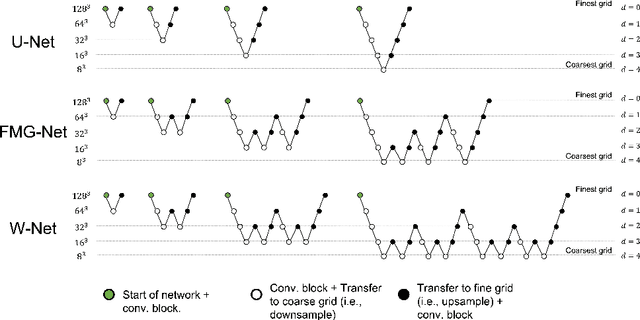
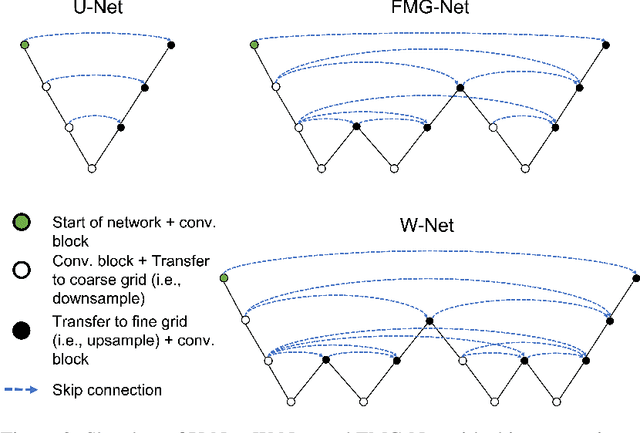
Abstract:Accurate medical imaging segmentation is critical for precise and effective medical interventions. However, despite the success of convolutional neural networks (CNNs) in medical image segmentation, they still face challenges in handling fine-scale features and variations in image scales. These challenges are particularly evident in complex and challenging segmentation tasks, such as the BraTS multi-label brain tumor segmentation challenge. In this task, accurately segmenting the various tumor sub-components, which vary significantly in size and shape, remains a significant challenge, with even state-of-the-art methods producing substantial errors. Therefore, we propose two architectures, FMG-Net and W-Net, that incorporate the principles of geometric multigrid methods for solving linear systems of equations into CNNs to address these challenges. Our experiments on the BraTS 2020 dataset demonstrate that both FMG-Net and W-Net outperform the widely used U-Net architecture regarding tumor subcomponent segmentation accuracy and training efficiency. These findings highlight the potential of incorporating the principles of multigrid methods into CNNs to improve the accuracy and efficiency of medical imaging segmentation.
A Weighted Normalized Boundary Loss for Reducing the Hausdorff Distance in Medical Imaging Segmentation
Feb 08, 2023Abstract:Within medical imaging segmentation, the Dice coefficient and Hausdorff-based metrics are standard measures of success for deep learning models. However, modern loss functions for medical image segmentation often only consider the Dice coefficient or similar region-based metrics during training. As a result, segmentation architectures trained over such loss functions run the risk of achieving high accuracy for the Dice coefficient but low accuracy for Hausdorff-based metrics. Low accuracy on Hausdorff-based metrics can be problematic for applications such as tumor segmentation, where such benchmarks are crucial. For example, high Dice scores accompanied by significant Hausdorff errors could indicate that the predictions fail to detect small tumors. We propose the Weighted Normalized Boundary Loss, a novel loss function to minimize Hausdorff-based metrics with more desirable numerical properties than current methods and with weighting terms for class imbalance. Our loss function outperforms other losses when tested on the BraTS dataset using a standard 3D U-Net and the state-of-the-art nnUNet architectures. These results suggest we can improve segmentation accuracy with our novel loss function.
Automation of Radiation Treatment Planning for Rectal Cancer
Apr 26, 2022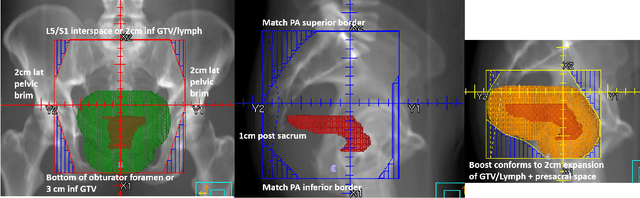
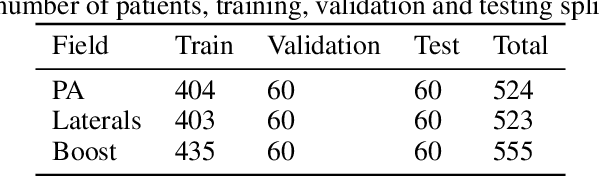
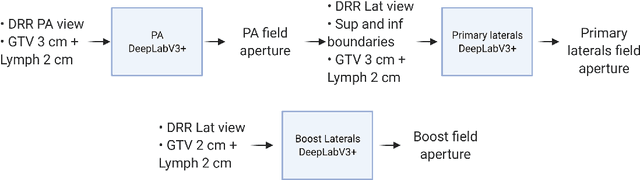
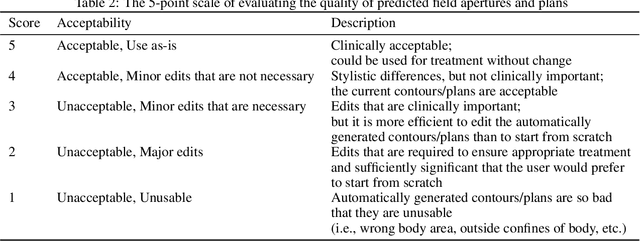
Abstract:To develop an automated workflow for rectal cancer three-dimensional conformal radiotherapy treatment planning that combines deep-learning(DL) aperture predictions and forward-planning algorithms. We designed an algorithm to automate the clinical workflow for planning with field-in-field. DL models were trained, validated, and tested on 555 patients to automatically generate aperture shapes for primary and boost fields. Network inputs were digitally reconstructed radiography, gross tumor volume(GTV), and nodal GTV. A physician scored each aperture for 20 patients on a 5-point scale(>3 acceptable). A planning algorithm was then developed to create a homogeneous dose using a combination of wedges and subfields. The algorithm iteratively identifies a hotspot volume, creates a subfield, and optimizes beam weight all without user intervention. The algorithm was tested on 20 patients using clinical apertures with different settings, and the resulting plans(4 plans/patient) were scored by a physician. The end-to-end workflow was tested and scored by a physician on 39 patients using DL-generated apertures and planning algorithms. The predicted apertures had Dice scores of 0.95, 0.94, and 0.90 for posterior-anterior, laterals, and boost fields, respectively. 100%, 95%, and 87.5% of the posterior-anterior, laterals, and boost apertures were scored as clinically acceptable, respectively. Wedged and non-wedged plans were clinically acceptable for 85% and 50% of patients, respectively. The final plans hotspot dose percentage was reduced from 121%($\pm$ 14%) to 109%($\pm$ 5%) of prescription dose. The integrated end-to-end workflow of automatically generated apertures and optimized field-in-field planning gave clinically acceptable plans for 38/39(97%) of patients. We have successfully automated the clinical workflow for generating radiotherapy plans for rectal cancer for our institution.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge