Rachel Glenn
Two Stage Segmentation of Cervical Tumors using PocketNet
Sep 17, 2024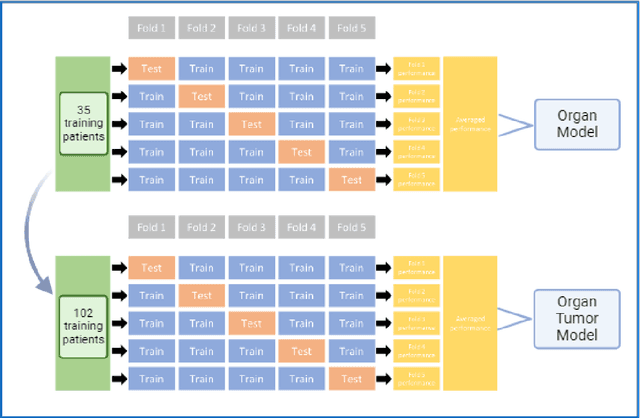

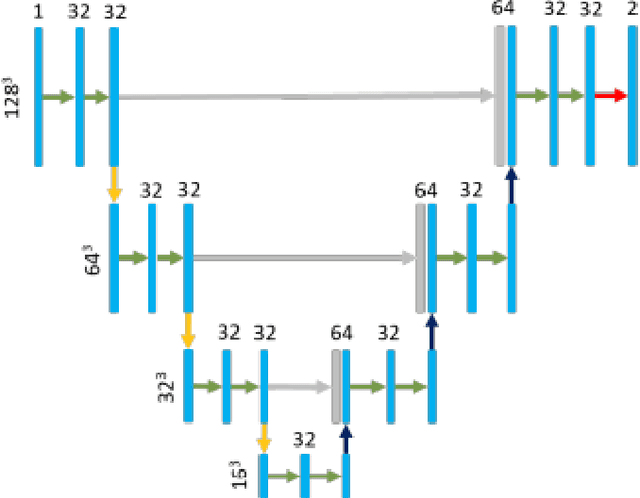
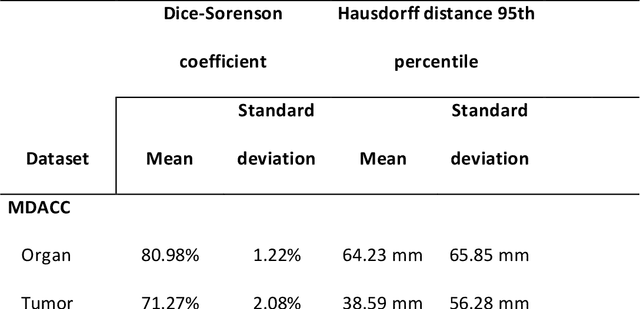
Abstract:Cervical cancer remains the fourth most common malignancy amongst women worldwide.1 Concurrent chemoradiotherapy (CRT) serves as the mainstay definitive treatment regimen for locally advanced cervical cancers and includes external beam radiation followed by brachytherapy.2 Integral to radiotherapy treatment planning is the routine contouring of both the target tumor at the level of the cervix, associated gynecologic anatomy and the adjacent organs at risk (OARs). However, manual contouring of these structures is both time and labor intensive and associated with known interobserver variability that can impact treatment outcomes. While multiple tools have been developed to automatically segment OARs and the high-risk clinical tumor volume (HR-CTV) using computed tomography (CT) images,3,4,5,6 the development of deep learning-based tumor segmentation tools using routine T2-weighted (T2w) magnetic resonance imaging (MRI) addresses an unmet clinical need to improve the routine contouring of both anatomical structures and cervical cancers, thereby increasing quality and consistency of radiotherapy planning. This work applied a novel deep-learning model (PocketNet) to segment the cervix, vagina, uterus, and tumor(s) on T2w MRI. The performance of the PocketNet architecture was evaluated, when trained on data via 5-fold cross validation. PocketNet achieved a mean Dice-Sorensen similarity coefficient (DSC) exceeding 70% for tumor segmentation and 80% for organ segmentation. These results suggest that PocketNet is robust to variations in contrast protocols, providing reliable segmentation of the ROIs.
MIST: A Simple and Scalable End-To-End 3D Medical Imaging Segmentation Framework
Jul 31, 2024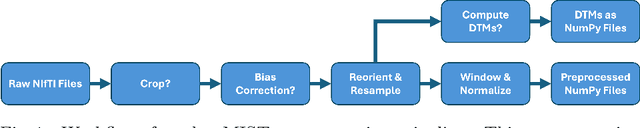
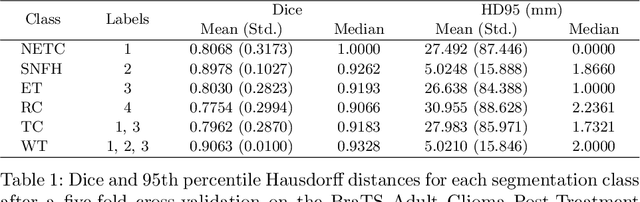
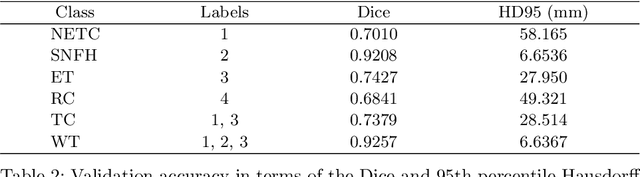
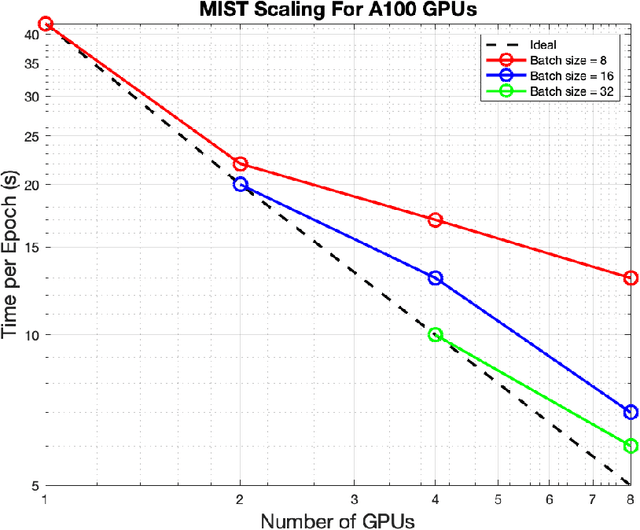
Abstract:Medical imaging segmentation is a highly active area of research, with deep learning-based methods achieving state-of-the-art results in several benchmarks. However, the lack of standardized tools for training, testing, and evaluating new methods makes the comparison of methods difficult. To address this, we introduce the Medical Imaging Segmentation Toolkit (MIST), a simple, modular, and end-to-end medical imaging segmentation framework designed to facilitate consistent training, testing, and evaluation of deep learning-based medical imaging segmentation methods. MIST standardizes data analysis, preprocessing, and evaluation pipelines, accommodating multiple architectures and loss functions. This standardization ensures reproducible and fair comparisons across different methods. We detail MIST's data format requirements, pipelines, and auxiliary features and demonstrate its efficacy using the BraTS Adult Glioma Post-Treatment Challenge dataset. Our results highlight MIST's ability to produce accurate segmentation masks and its scalability across multiple GPUs, showcasing its potential as a powerful tool for future medical imaging research and development.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge