Alex Balsells
MIST: A Simple and Scalable End-To-End 3D Medical Imaging Segmentation Framework
Jul 31, 2024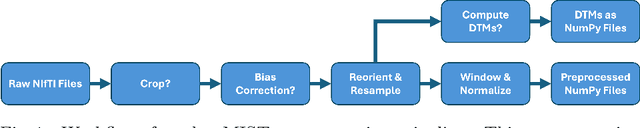
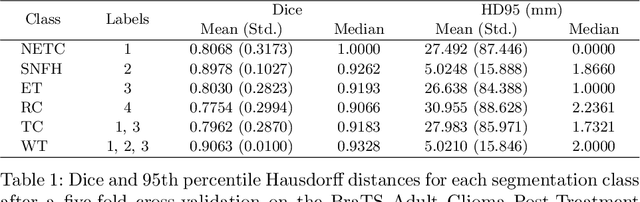
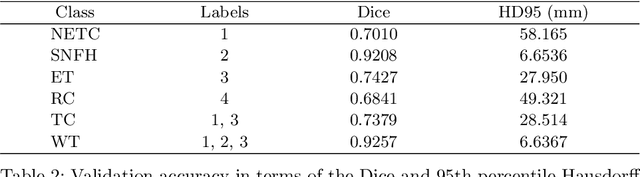
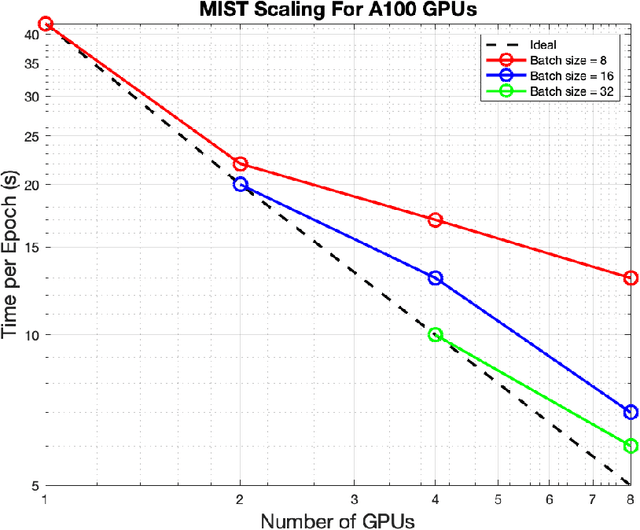
Abstract:Medical imaging segmentation is a highly active area of research, with deep learning-based methods achieving state-of-the-art results in several benchmarks. However, the lack of standardized tools for training, testing, and evaluating new methods makes the comparison of methods difficult. To address this, we introduce the Medical Imaging Segmentation Toolkit (MIST), a simple, modular, and end-to-end medical imaging segmentation framework designed to facilitate consistent training, testing, and evaluation of deep learning-based medical imaging segmentation methods. MIST standardizes data analysis, preprocessing, and evaluation pipelines, accommodating multiple architectures and loss functions. This standardization ensures reproducible and fair comparisons across different methods. We detail MIST's data format requirements, pipelines, and auxiliary features and demonstrate its efficacy using the BraTS Adult Glioma Post-Treatment Challenge dataset. Our results highlight MIST's ability to produce accurate segmentation masks and its scalability across multiple GPUs, showcasing its potential as a powerful tool for future medical imaging research and development.
A Weighted Normalized Boundary Loss for Reducing the Hausdorff Distance in Medical Imaging Segmentation
Feb 08, 2023Abstract:Within medical imaging segmentation, the Dice coefficient and Hausdorff-based metrics are standard measures of success for deep learning models. However, modern loss functions for medical image segmentation often only consider the Dice coefficient or similar region-based metrics during training. As a result, segmentation architectures trained over such loss functions run the risk of achieving high accuracy for the Dice coefficient but low accuracy for Hausdorff-based metrics. Low accuracy on Hausdorff-based metrics can be problematic for applications such as tumor segmentation, where such benchmarks are crucial. For example, high Dice scores accompanied by significant Hausdorff errors could indicate that the predictions fail to detect small tumors. We propose the Weighted Normalized Boundary Loss, a novel loss function to minimize Hausdorff-based metrics with more desirable numerical properties than current methods and with weighting terms for class imbalance. Our loss function outperforms other losses when tested on the BraTS dataset using a standard 3D U-Net and the state-of-the-art nnUNet architectures. These results suggest we can improve segmentation accuracy with our novel loss function.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge