Caihong Zeng
Unsupervised Tissue Segmentation via Deep Constrained Gaussian Network
Aug 04, 2022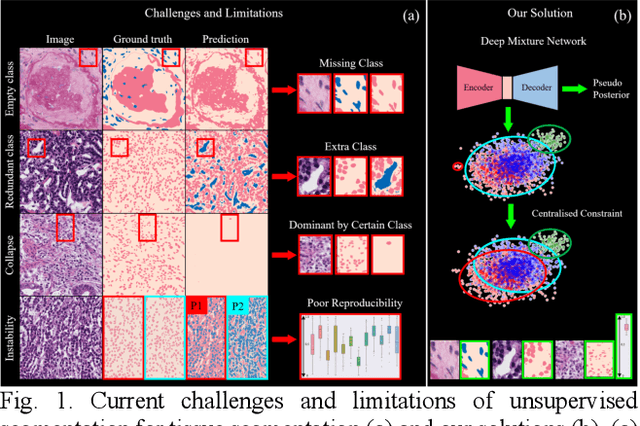
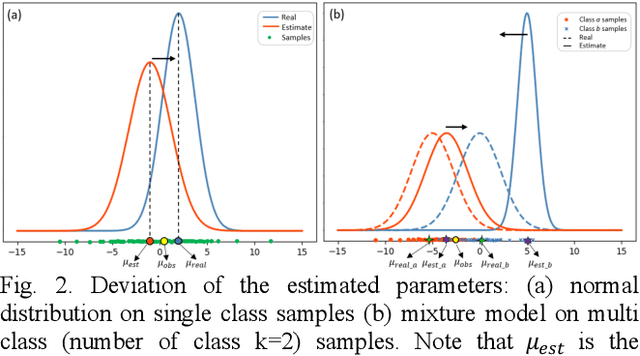
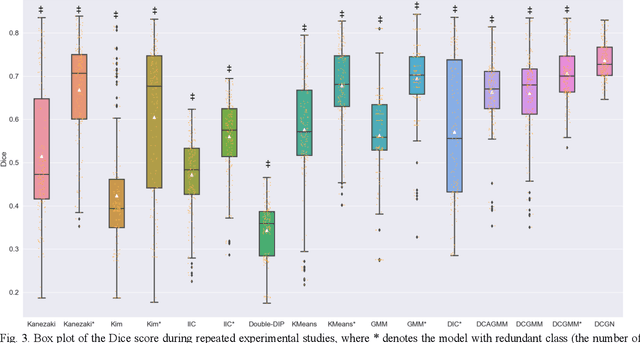
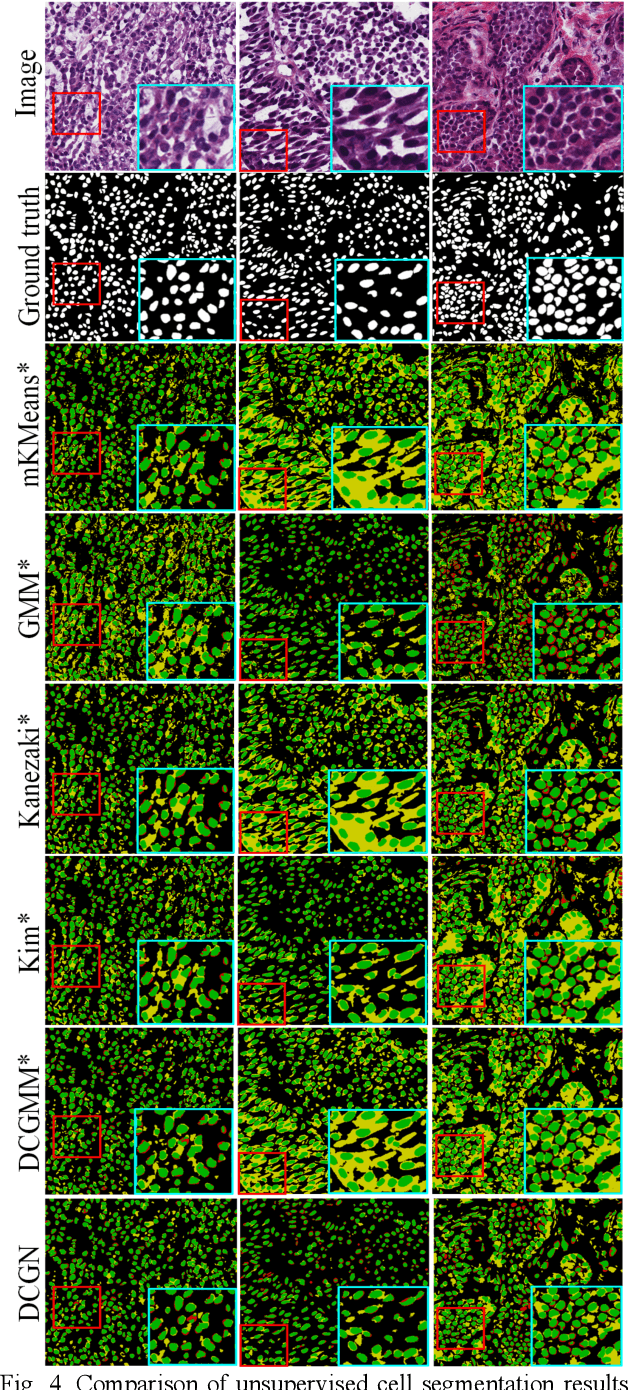
Abstract:Tissue segmentation is the mainstay of pathological examination, whereas the manual delineation is unduly burdensome. To assist this time-consuming and subjective manual step, researchers have devised methods to automatically segment structures in pathological images. Recently, automated machine and deep learning based methods dominate tissue segmentation research studies. However, most machine and deep learning based approaches are supervised and developed using a large number of training samples, in which the pixelwise annotations are expensive and sometimes can be impossible to obtain. This paper introduces a novel unsupervised learning paradigm by integrating an end-to-end deep mixture model with a constrained indicator to acquire accurate semantic tissue segmentation. This constraint aims to centralise the components of deep mixture models during the calculation of the optimisation function. In so doing, the redundant or empty class issues, which are common in current unsupervised learning methods, can be greatly reduced. By validation on both public and in-house datasets, the proposed deep constrained Gaussian network achieves significantly (Wilcoxon signed-rank test) better performance (with the average Dice scores of 0.737 and 0.735, respectively) on tissue segmentation with improved stability and robustness, compared to other existing unsupervised segmentation approaches. Furthermore, the proposed method presents a similar performance (p-value > 0.05) compared to the fully supervised U-Net.
Automatic Fine-grained Glomerular Lesion Recognition in Kidney Pathology
Mar 11, 2022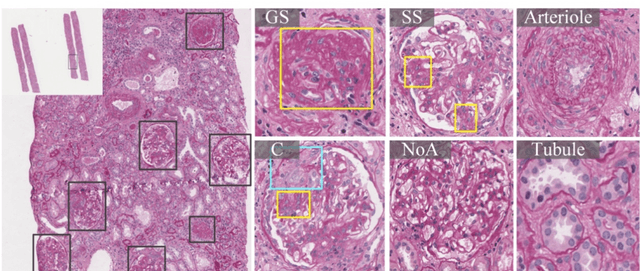
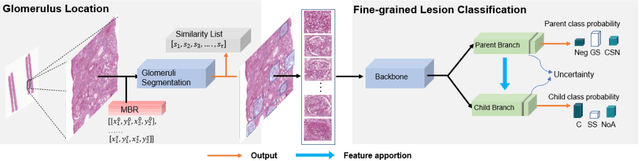
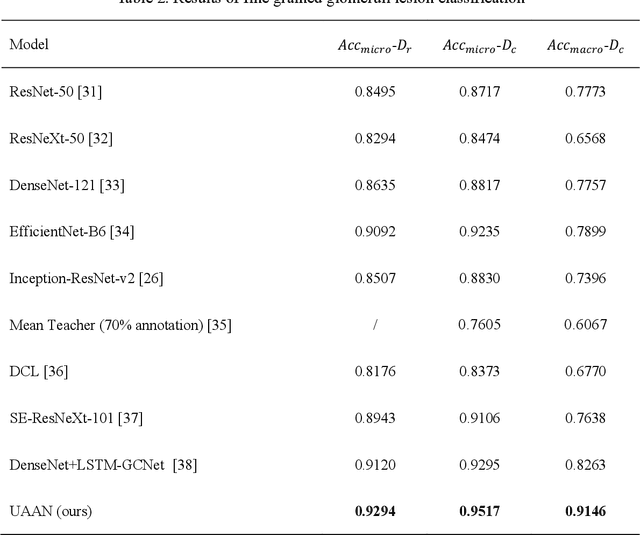
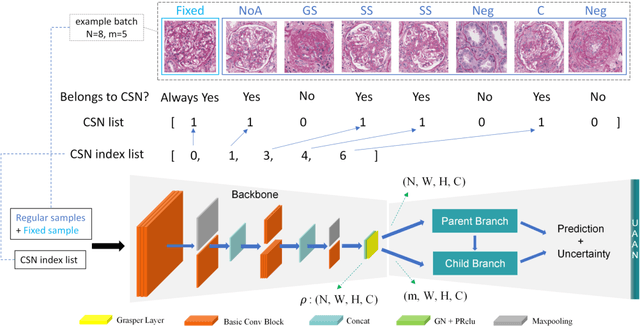
Abstract:Recognition of glomeruli lesions is the key for diagnosis and treatment planning in kidney pathology; however, the coexisting glomerular structures such as mesangial regions exacerbate the difficulties of this task. In this paper, we introduce a scheme to recognize fine-grained glomeruli lesions from whole slide images. First, a focal instance structural similarity loss is proposed to drive the model to locate all types of glomeruli precisely. Then an Uncertainty Aided Apportionment Network is designed to carry out the fine-grained visual classification without bounding-box annotations. This double branch-shaped structure extracts common features of the child class from the parent class and produces the uncertainty factor for reconstituting the training dataset. Results of slide-wise evaluation illustrate the effectiveness of the entire scheme, with an 8-22% improvement of the mean Average Precision compared with remarkable detection methods. The comprehensive results clearly demonstrate the effectiveness of the proposed method.
P3SGD: Patient Privacy Preserving SGD for Regularizing Deep CNNs in Pathological Image Classification
May 30, 2019
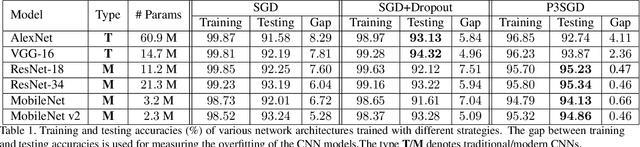
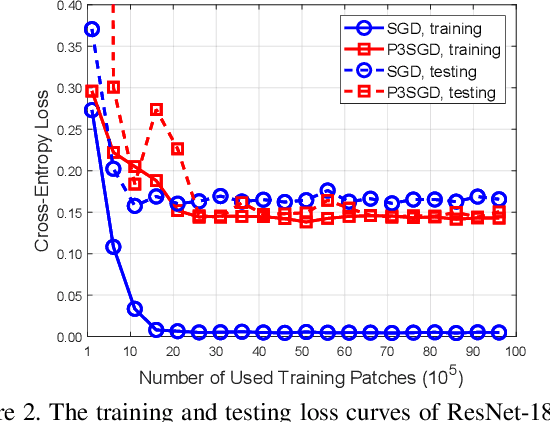
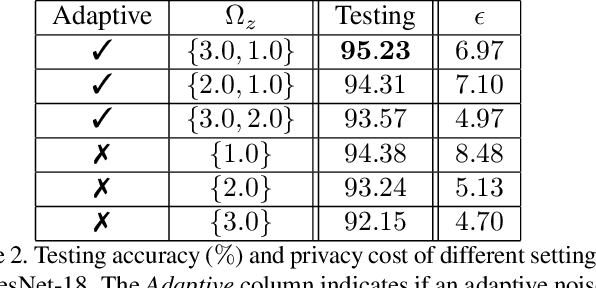
Abstract:Recently, deep convolutional neural networks (CNNs) have achieved great success in pathological image classification. However, due to the limited number of labeled pathological images, there are still two challenges to be addressed: (1) overfitting: the performance of a CNN model is undermined by the overfitting due to its huge amounts of parameters and the insufficiency of labeled training data. (2) privacy leakage: the model trained using a conventional method may involuntarily reveal the private information of the patients in the training dataset. The smaller the dataset, the worse the privacy leakage. To tackle the above two challenges, we introduce a novel stochastic gradient descent (SGD) scheme, named patient privacy preserving SGD (P3SGD), which performs the model update of the SGD in the patient level via a large-step update built upon each patient's data. Specifically, to protect privacy and regularize the CNN model, we propose to inject the well-designed noise into the updates. Moreover, we equip our P3SGD with an elaborated strategy to adaptively control the scale of the injected noise. To validate the effectiveness of P3SGD, we perform extensive experiments on a real-world clinical dataset and quantitatively demonstrate the superior ability of P3SGD in reducing the risk of overfitting. We also provide a rigorous analysis of the privacy cost under differential privacy. Additionally, we find that the models trained with P3SGD are resistant to the model-inversion attack compared with those trained using non-private SGD.
G2C: A Generator-to-Classifier Framework Integrating Multi-Stained Visual Cues for Pathological Glomerulus Classification
Jun 30, 2018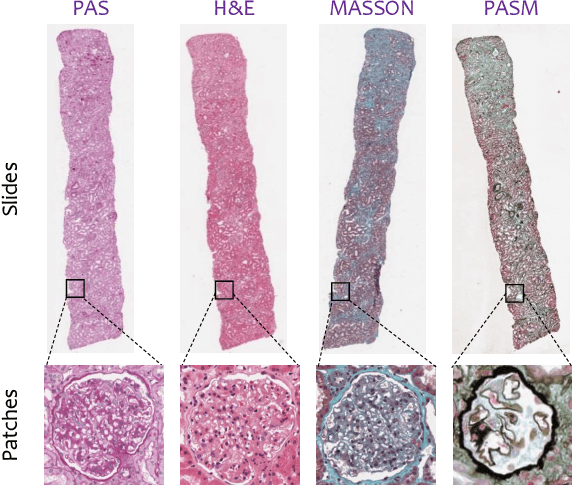
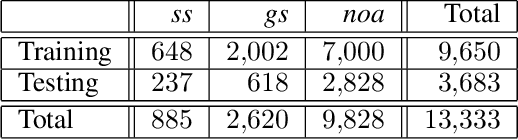
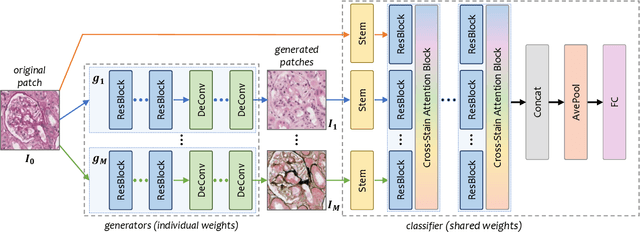
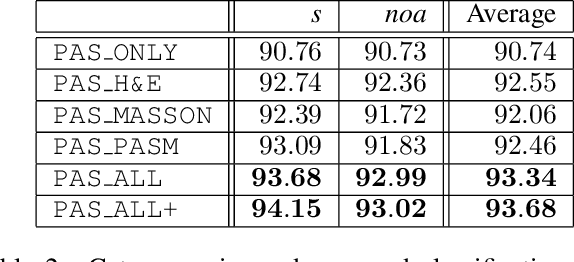
Abstract:Pathological glomerulus classification plays a key role in the diagnosis of nephropathy. As the difference between different subcategories is subtle, doctors often refer to slides from different staining methods to make decisions. However, creating correspondence across various stains is labor-intensive, bringing major difficulties in collecting data and training a vision-based algorithm to assist nephropathy diagnosis. This paper provides an alternative solution for integrating multi-stained visual cues for glomerulus classification. Our approach, named generator-to-classifier (G2C), is a two-stage framework. Given an input image from a specified stain, several generators are first applied to estimate its appearances in other staining methods, and a classifier follows to combine these visual cues for decision making. These two stages are optimized in a joint manner. To provide a reasonable initialization for the generators, we train an unpaired image-to-image translation network for each stain, and fine-tune them with the classifier. Since there are no publicly available datasets for glomerulus classification, we collect one by ourselves. Experiments reveal the effectiveness of our approach, including the authenticity of the generated patches so that doctors can hardly distinguish them from the real ones. We also transfer our model to a public dataset for breast cancer classification, and outperform the state-of-the-arts significantly
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge