Bolei Xu
Class-Aware Domain Adaptation for Improving Adversarial Robustness
May 10, 2020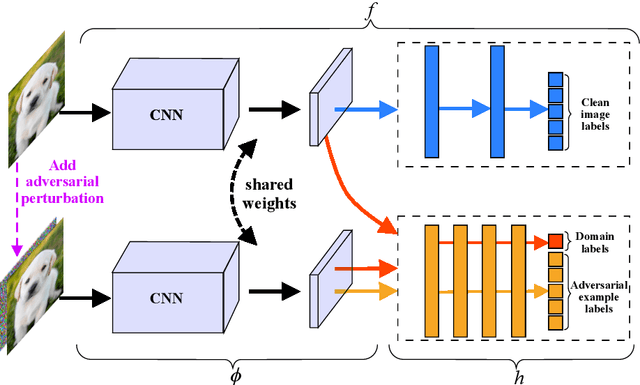
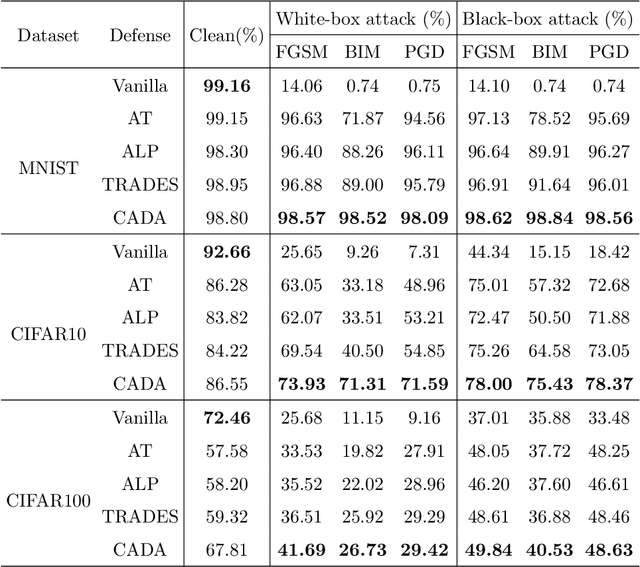
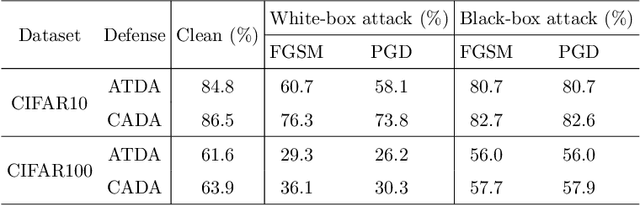
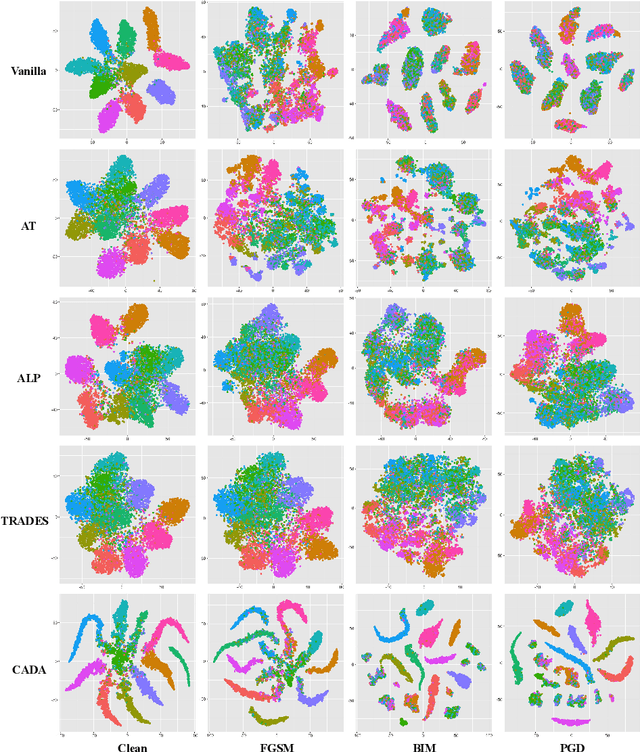
Abstract:Recent works have demonstrated convolutional neural networks are vulnerable to adversarial examples, i.e., inputs to machine learning models that an attacker has intentionally designed to cause the models to make a mistake. To improve the adversarial robustness of neural networks, adversarial training has been proposed to train networks by injecting adversarial examples into the training data. However, adversarial training could overfit to a specific type of adversarial attack and also lead to standard accuracy drop on clean images. To this end, we propose a novel Class-Aware Domain Adaptation (CADA) method for adversarial defense without directly applying adversarial training. Specifically, we propose to learn domain-invariant features for adversarial examples and clean images via a domain discriminator. Furthermore, we introduce a class-aware component into the discriminator to increase the discriminative power of the network for adversarial examples. We evaluate our newly proposed approach using multiple benchmark datasets. The results demonstrate that our method can significantly improve the state-of-the-art of adversarial robustness for various attacks and maintain high performances on clean images.
Dual Adaptive Pyramid Network for Cross-Stain Histopathology Image Segmentation
Sep 25, 2019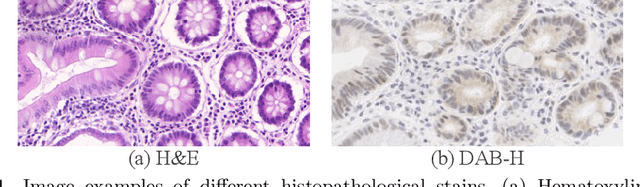
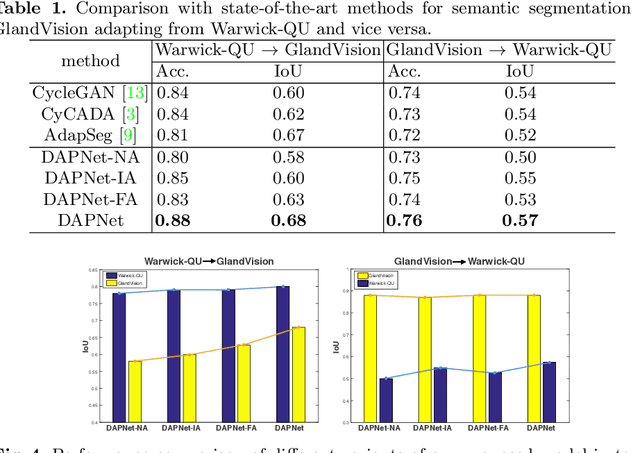
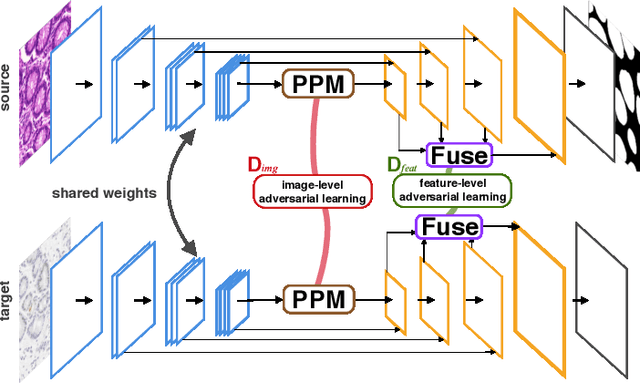
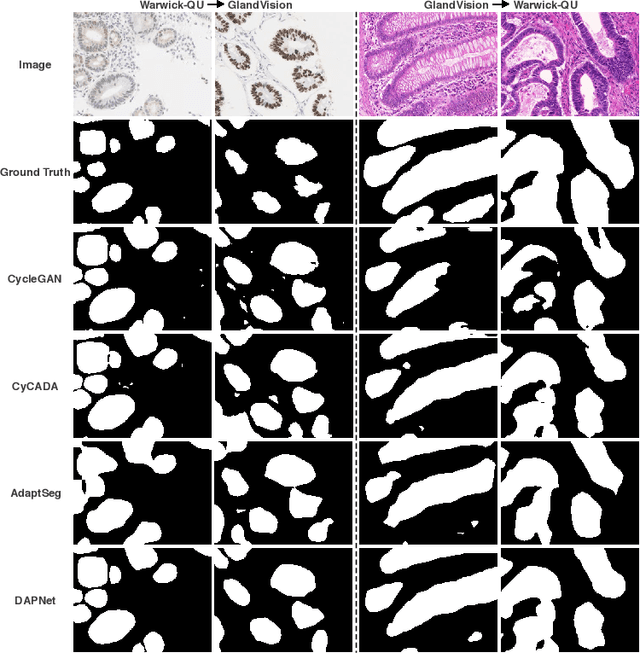
Abstract:Supervised semantic segmentation normally assumes the test data being in a similar data domain as the training data. However, in practice, the domain mismatch between the training and unseen data could lead to a significant performance drop. Obtaining accurate pixel-wise label for images in different domains is tedious and labor intensive, especially for histopathology images. In this paper, we propose a dual adaptive pyramid network (DAPNet) for histopathological gland segmentation adapting from one stain domain to another. We tackle the domain adaptation problem on two levels: 1) the image-level considers the differences of image color and style; 2) the feature-level addresses the spatial inconsistency between two domains. The two components are implemented as domain classifiers with adversarial training. We evaluate our new approach using two gland segmentation datasets with H&E and DAB-H stains respectively. The extensive experiments and ablation study demonstrate the effectiveness of our approach on the domain adaptive segmentation task. We show that the proposed approach performs favorably against other state-of-the-art methods.
Learning Deep Image Priors for Blind Image Denoising
Jun 04, 2019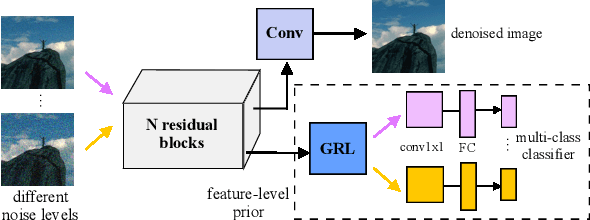
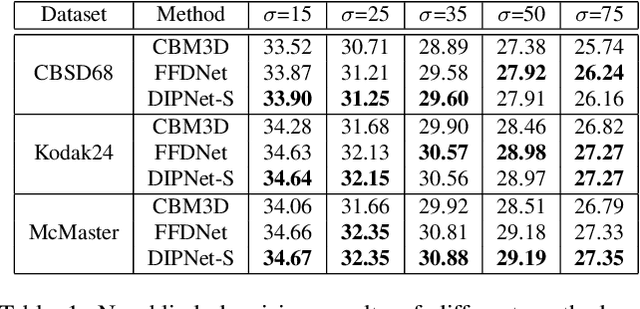
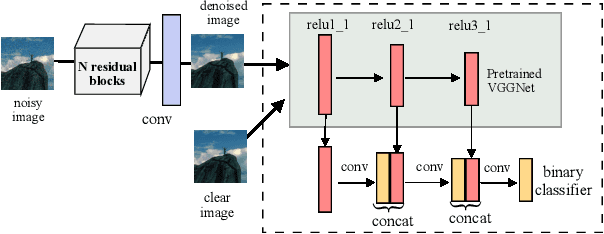
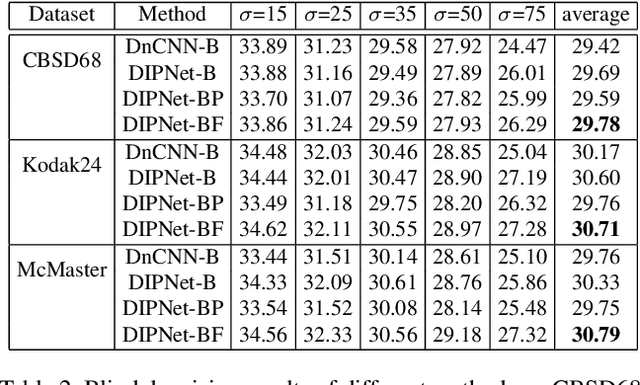
Abstract:Image denoising is the process of removing noise from noisy images, which is an image domain transferring task, i.e., from a single or several noise level domains to a photo-realistic domain. In this paper, we propose an effective image denoising method by learning two image priors from the perspective of domain alignment. We tackle the domain alignment on two levels. 1) the feature-level prior is to learn domain-invariant features for corrupted images with different level noise; 2) the pixel-level prior is used to push the denoised images to the natural image manifold. The two image priors are based on $\mathcal{H}$-divergence theory and implemented by learning classifiers in adversarial training manners. We evaluate our approach on multiple datasets. The results demonstrate the effectiveness of our approach for robust image denoising on both synthetic and real-world noisy images. Furthermore, we show that the feature-level prior is capable of alleviating the discrepancy between different level noise. It can be used to improve the blind denoising performance in terms of distortion measures (PSNR and SSIM), while pixel-level prior can effectively improve the perceptual quality to ensure the realistic outputs, which is further validated by subjective evaluation.
Look, Investigate, and Classify: A Deep Hybrid Attention Method for Breast Cancer Classification
Feb 28, 2019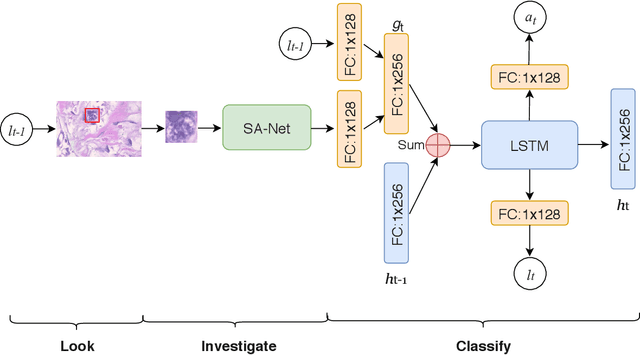
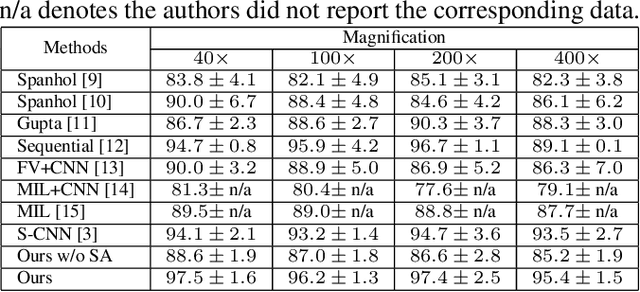
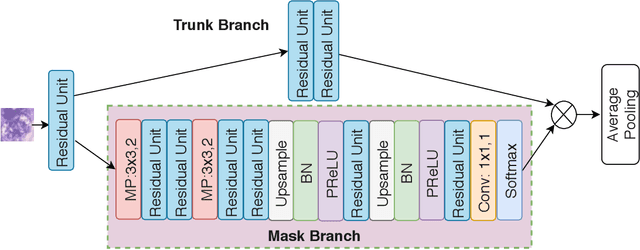
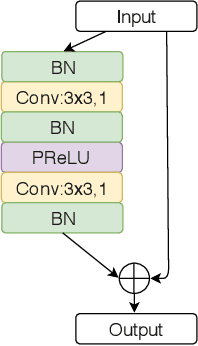
Abstract:One issue with computer based histopathology image analysis is that the size of the raw image is usually very large. Taking the raw image as input to the deep learning model would be computationally expensive while resizing the raw image to low resolution would incur information loss. In this paper, we present a novel deep hybrid attention approach to breast cancer classification. It first adaptively selects a sequence of coarse regions from the raw image by a hard visual attention algorithm, and then for each such region it is able to investigate the abnormal parts based on a soft-attention mechanism. A recurrent network is then built to make decisions to classify the image region and also to predict the location of the image region to be investigated at the next time step. As the region selection process is non-differentiable, we optimize the whole network through a reinforcement approach to learn an optimal policy to classify the regions. Based on this novel Look, Investigate and Classify approach, we only need to process a fraction of the pixels in the raw image resulting in significant saving in computational resources without sacrificing performances. Our approach is evaluated on a public breast cancer histopathology database, where it demonstrates superior performance to the state-of-the-art deep learning approaches, achieving around 96\% classification accuracy while only 15% of raw pixels are used.
An End-to-End Deep Learning Histochemical Scoring System for Breast Cancer Tissue Microarray
Jan 19, 2018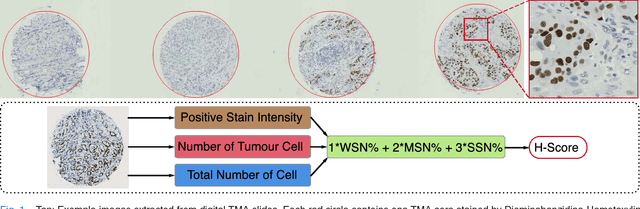
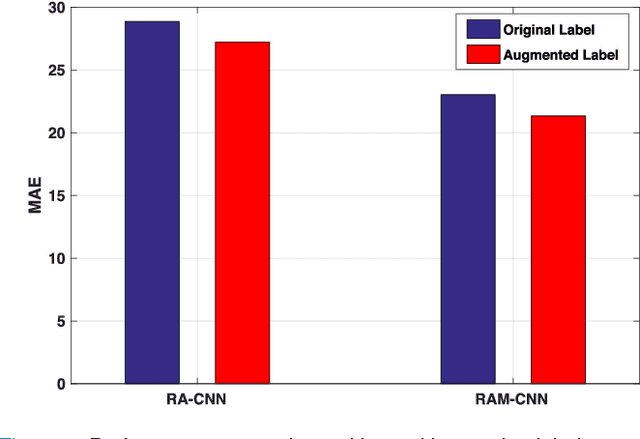
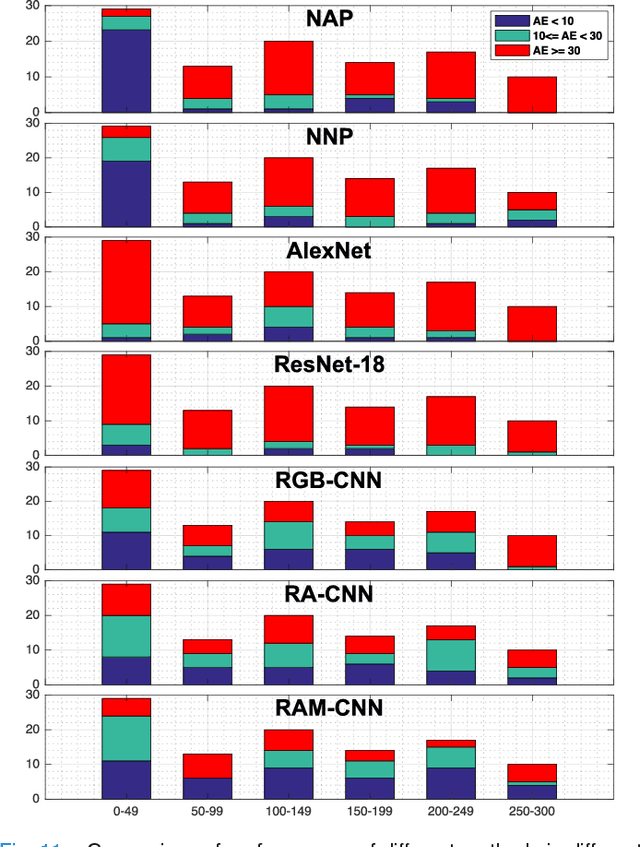
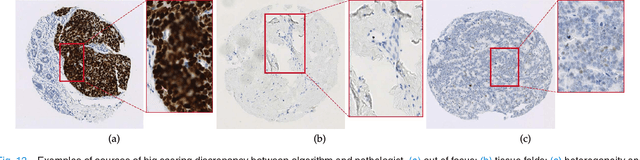
Abstract:One of the methods for stratifying different molecular classes of breast cancer is the Nottingham Prognostic Index Plus (NPI+) which uses breast cancer relevant biomarkers to stain tumour tissues prepared on tissue microarray (TMA). To determine the molecular class of the tumour, pathologists will have to manually mark the nuclei activity biomarkers through a microscope and use a semi-quantitative assessment method to assign a histochemical score (H-Score) to each TMA core. Manually marking positively stained nuclei is a time consuming, imprecise and subjective process which will lead to inter-observer and intra-observer discrepancies. In this paper, we present an end-to-end deep learning system which directly predicts the H-Score automatically. Our system imitates the pathologists' decision process and uses one fully convolutional network (FCN) to extract all nuclei region (tumour and non-tumour), a second FCN to extract tumour nuclei region, and a multi-column convolutional neural network which takes the outputs of the first two FCNs and the stain intensity description image as input and acts as the high-level decision making mechanism to directly output the H-Score of the input TMA image. To the best of our knowledge, this is the first end-to-end system that takes a TMA image as input and directly outputs a clinical score. We will present experimental results which demonstrate that the H-Scores predicted by our model have very high and statistically significant correlation with experienced pathologists' scores and that the H-Score discrepancy between our algorithm and the pathologists is on par with the inter-subject discrepancy between the pathologists.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge