Mohammad Ilyas
A Hybrid CNN-LSTM Deep Learning Model for Intrusion Detection in Smart Grid
Sep 08, 2025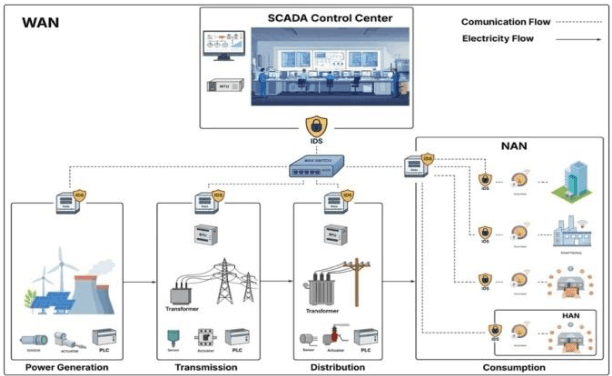

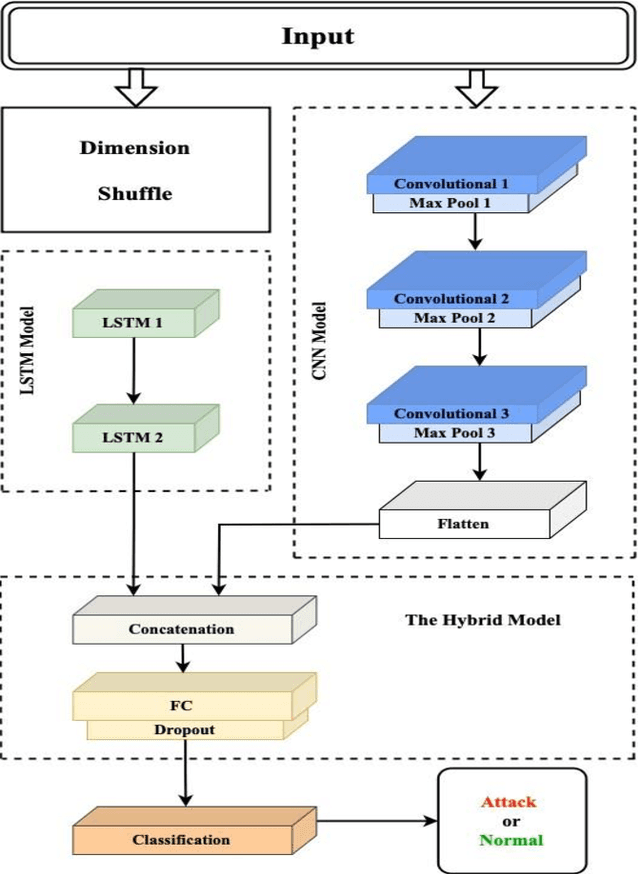

Abstract:The evolution of the traditional power grid into the "smart grid" has resulted in a fundamental shift in energy management, which allows the integration of renewable energy sources with modern communication technology. However, this interconnection has increased smart grids' vulnerability to attackers, which might result in privacy breaches, operational interruptions, and massive outages. The SCADA-based smart grid protocols are critical for real-time data collection and control, but they are vulnerable to attacks like unauthorized access and denial of service (DoS). This research proposes a hybrid deep learning-based Intrusion Detection System (IDS) intended to improve the cybersecurity of smart grids. The suggested model takes advantage of Convolutional Neural Networks' (CNN) feature extraction capabilities as well as Long Short-Term Memory (LSTM) networks' temporal pattern recognition skills. DNP3 and IEC104 intrusion detection datasets are employed to train and test our CNN-LSTM model to recognize and classify the potential cyber threats. Compared to other deep learning approaches, the results demonstrate considerable improvements in accuracy, precision, recall, and F1-score, with a detection accuracy of 99.70%.
A Machine Learning Ensemble Model for the Detection of Cyberbullying
Feb 19, 2024Abstract:The pervasive use of social media platforms, such as Facebook, Instagram, and X, has significantly amplified our electronic interconnectedness. Moreover, these platforms are now easily accessible from any location at any given time. However, the increased popularity of social media has also led to cyberbullying.It is imperative to address the need for finding, monitoring, and mitigating cyberbullying posts on social media platforms. Motivated by this necessity, we present this paper to contribute to developing an automated system for detecting binary labels of aggressive tweets.Our study has demonstrated remarkable performance compared to previous experiments on the same dataset. We employed the stacking ensemble machine learning method, utilizing four various feature extraction techniques to optimize performance within the stacking ensemble learning framework. Combining five machine learning algorithms,Decision Trees, Random Forest, Linear Support Vector Classification, Logistic Regression, and K-Nearest Neighbors into an ensemble method, we achieved superior results compared to traditional machine learning classifier models. The stacking classifier achieved a high accuracy rate of 94.00%, outperforming traditional machine learning models and surpassing the results of prior experiments that utilized the same dataset. The outcomes of our experiments showcased an accuracy rate of 0.94% in detection tweets as aggressive or non-aggressive.
Dual Adaptive Pyramid Network for Cross-Stain Histopathology Image Segmentation
Sep 25, 2019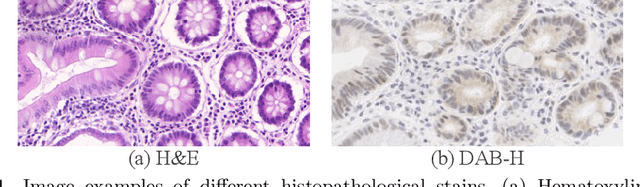
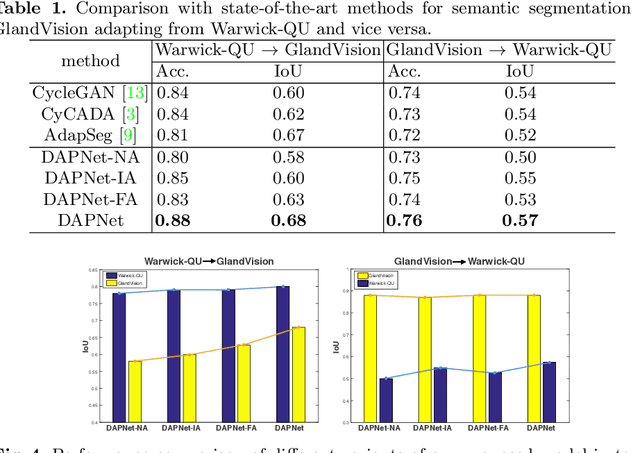
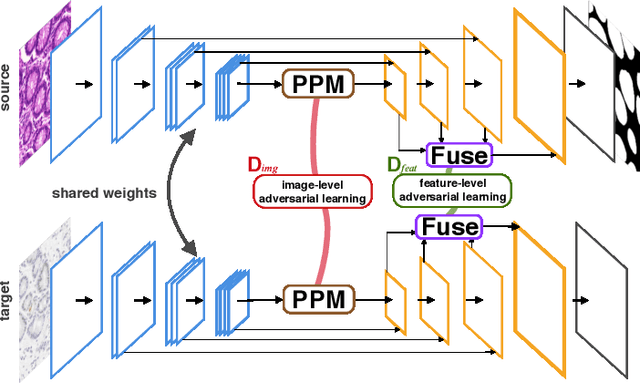
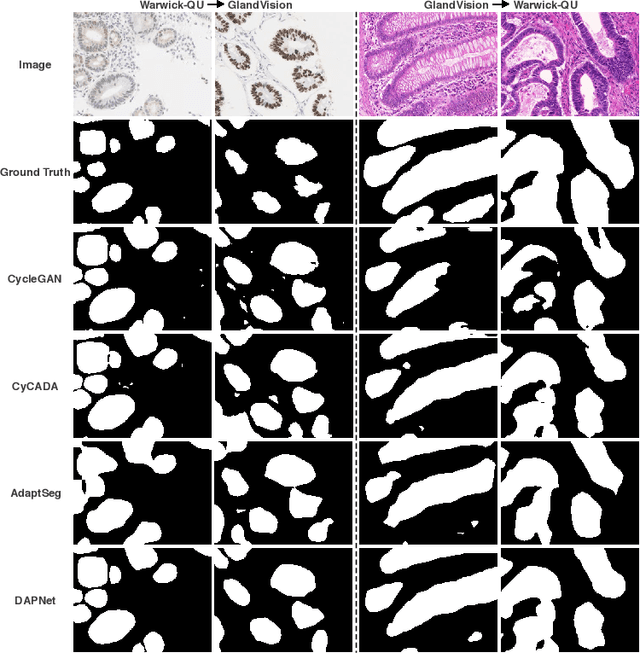
Abstract:Supervised semantic segmentation normally assumes the test data being in a similar data domain as the training data. However, in practice, the domain mismatch between the training and unseen data could lead to a significant performance drop. Obtaining accurate pixel-wise label for images in different domains is tedious and labor intensive, especially for histopathology images. In this paper, we propose a dual adaptive pyramid network (DAPNet) for histopathological gland segmentation adapting from one stain domain to another. We tackle the domain adaptation problem on two levels: 1) the image-level considers the differences of image color and style; 2) the feature-level addresses the spatial inconsistency between two domains. The two components are implemented as domain classifiers with adversarial training. We evaluate our new approach using two gland segmentation datasets with H&E and DAB-H stains respectively. The extensive experiments and ablation study demonstrate the effectiveness of our approach on the domain adaptive segmentation task. We show that the proposed approach performs favorably against other state-of-the-art methods.
Her2 Challenge Contest: A Detailed Assessment of Automated Her2 Scoring Algorithms in Whole Slide Images of Breast Cancer Tissues
Jul 24, 2017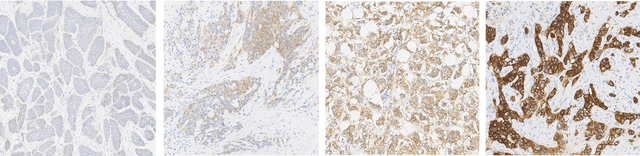
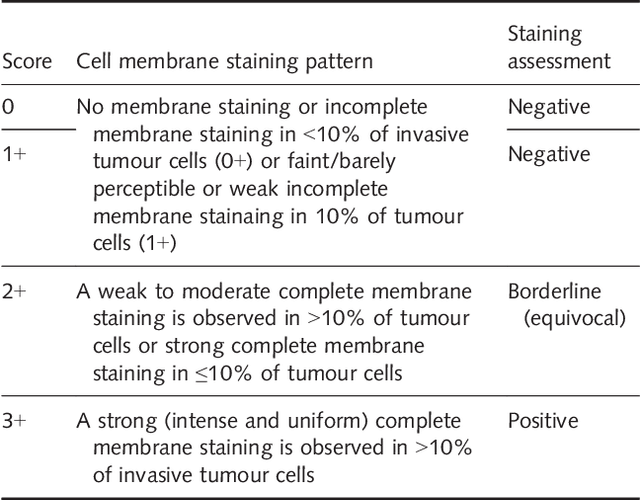
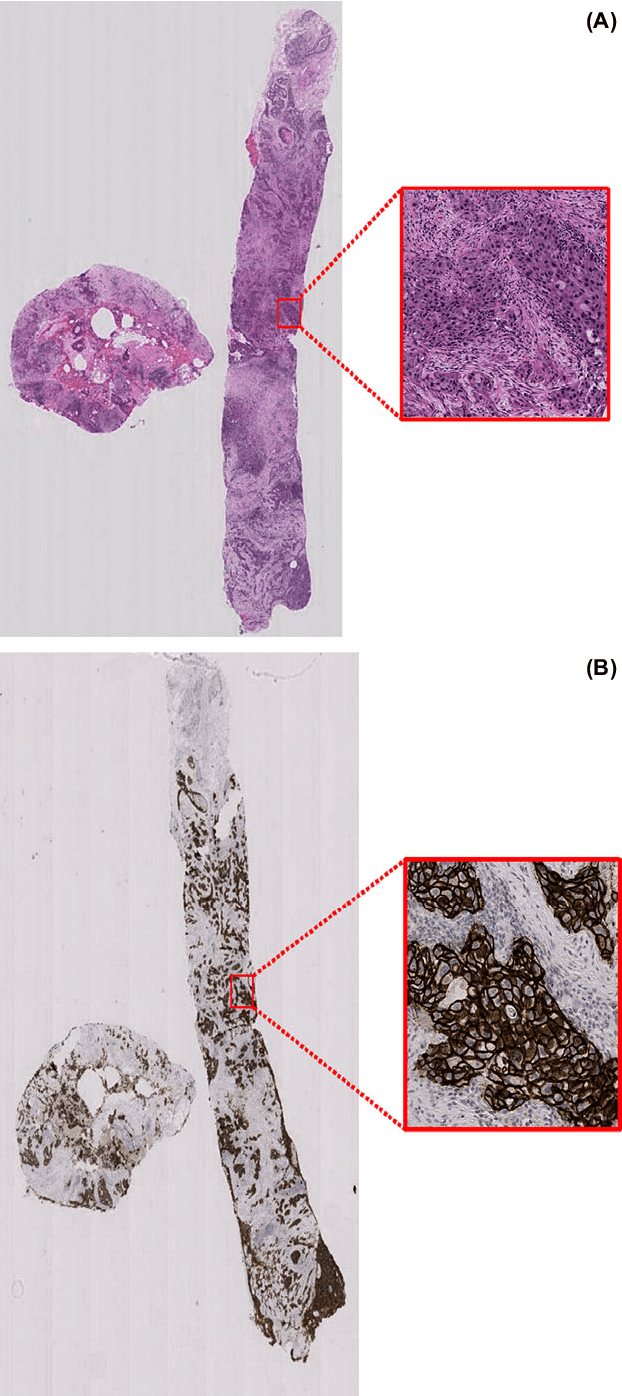
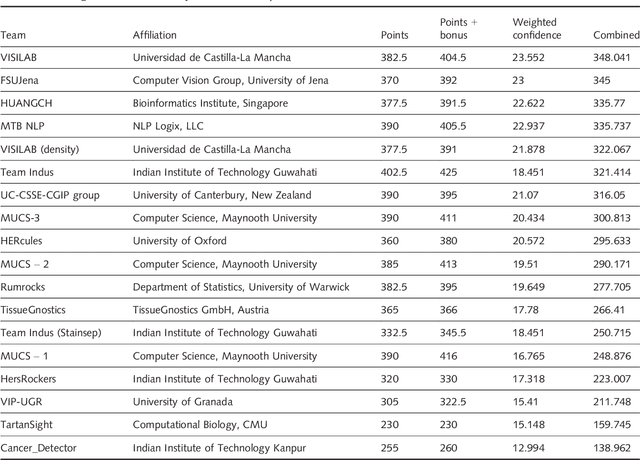
Abstract:Evaluating expression of the Human epidermal growth factor receptor 2 (Her2) by visual examination of immunohistochemistry (IHC) on invasive breast cancer (BCa) is a key part of the diagnostic assessment of BCa due to its recognised importance as a predictive and prognostic marker in clinical practice. However, visual scoring of Her2 is subjective and consequently prone to inter-observer variability. Given the prognostic and therapeutic implications of Her2 scoring, a more objective method is required. In this paper, we report on a recent automated Her2 scoring contest, held in conjunction with the annual PathSoc meeting held in Nottingham in June 2016, aimed at systematically comparing and advancing the state-of-the-art Artificial Intelligence (AI) based automated methods for Her2 scoring. The contest dataset comprised of digitised whole slide images (WSI) of sections from 86 cases of invasive breast carcinoma stained with both Haematoxylin & Eosin (H&E) and IHC for Her2. The contesting algorithms automatically predicted scores of the IHC slides for an unseen subset of the dataset and the predicted scores were compared with the 'ground truth' (a consensus score from at least two experts). We also report on a simple Man vs Machine contest for the scoring of Her2 and show that the automated methods could beat the pathology experts on this contest dataset. This paper presents a benchmark for comparing the performance of automated algorithms for scoring of Her2. It also demonstrates the enormous potential of automated algorithms in assisting the pathologist with objective IHC scoring.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge