Bogdan Sacaleanu
CHARD: Clinical Health-Aware Reasoning Across Dimensions for Text Generation Models
Oct 09, 2022
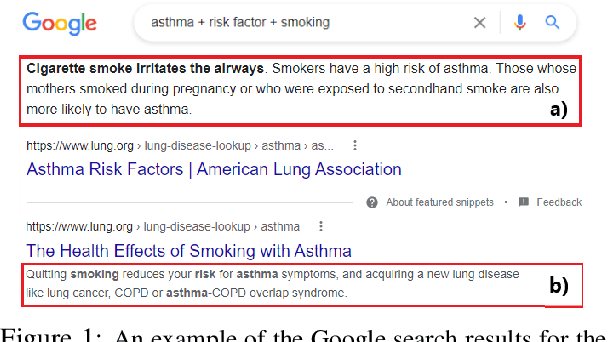
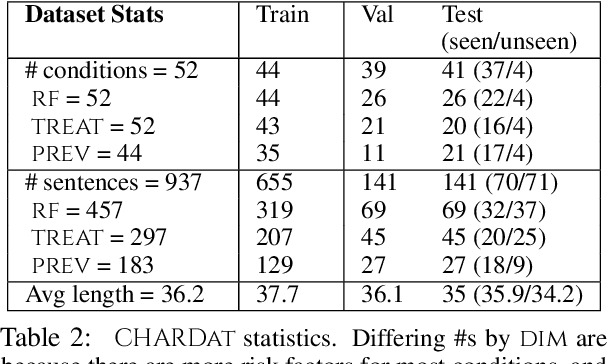
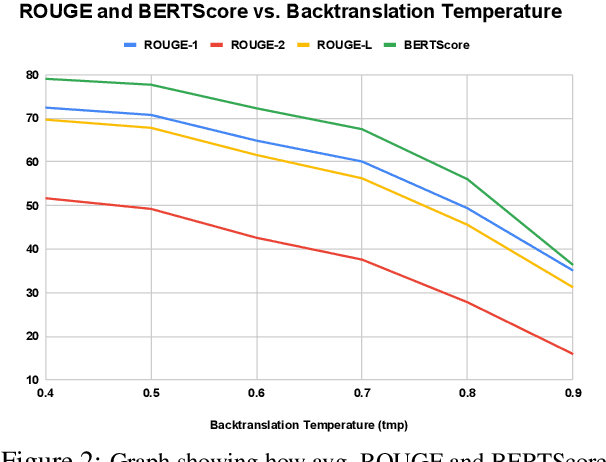
Abstract:We motivate and introduce CHARD: Clinical Health-Aware Reasoning across Dimensions, to investigate the capability of text generation models to act as implicit clinical knowledge bases and generate free-flow textual explanations about various health-related conditions across several dimensions. We collect and present an associated dataset, CHARDat, consisting of explanations about 52 health conditions across three clinical dimensions. We conduct extensive experiments using BART and T5 along with data augmentation, and perform automatic, human, and qualitative analyses. We show that while our models can perform decently, CHARD is very challenging with strong potential for further exploration.
Cross-Domain Reasoning via Template Filling
Oct 31, 2021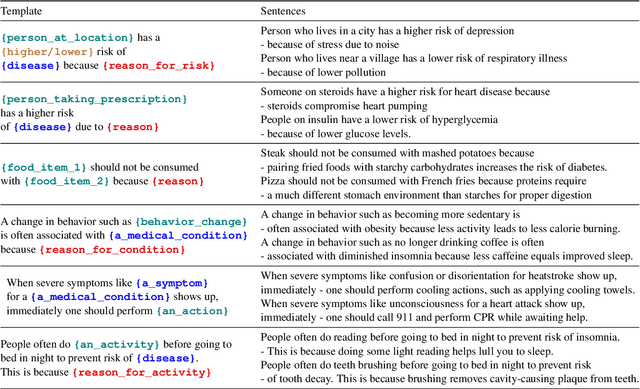
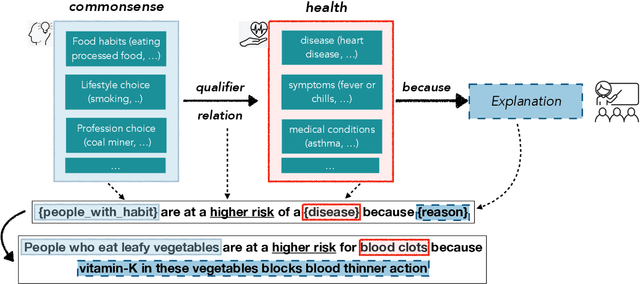
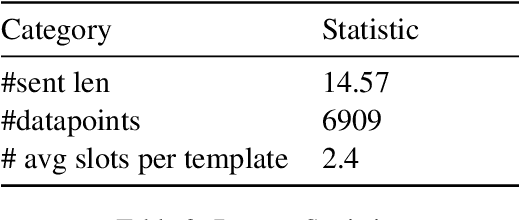

Abstract:In this paper, we explore the ability of sequence to sequence models to perform cross-domain reasoning. Towards this, we present a prompt-template-filling approach to enable sequence to sequence models to perform cross-domain reasoning. We also present a case-study with commonsense and health and well-being domains, where we study how prompt-template-filling enables pretrained sequence to sequence models across domains. Our experiments across several pretrained encoder-decoder models show that cross-domain reasoning is challenging for current models. We also show an in-depth error analysis and avenues for future research for reasoning across domains
MIMICause : Defining, identifying and predicting types of causal relationships between biomedical concepts from clinical notes
Oct 14, 2021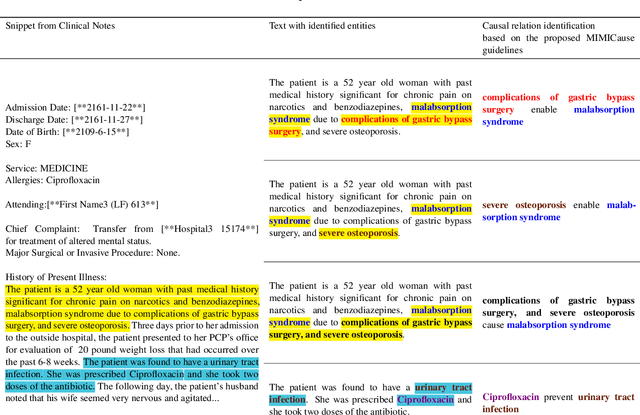
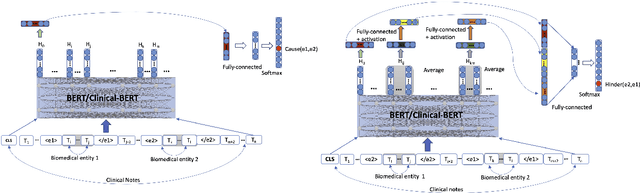
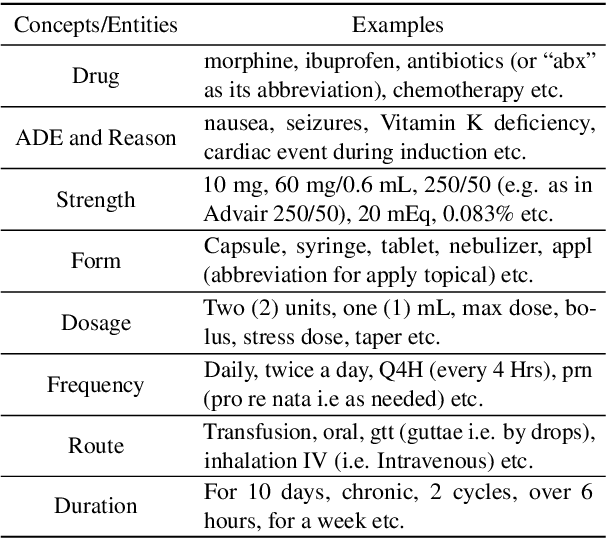
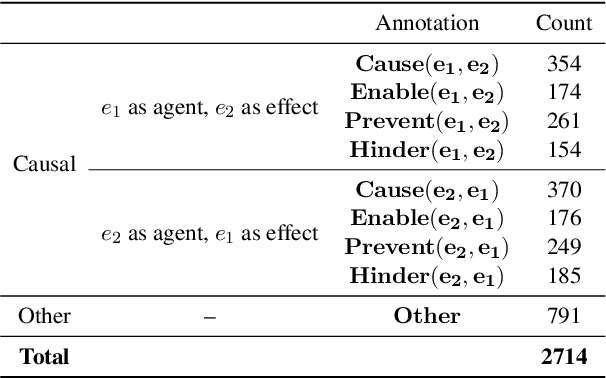
Abstract:Understanding of causal narratives communicated in clinical notes can help make strides towards personalized healthcare. In this work, MIMICause, we propose annotation guidelines, develop an annotated corpus and provide baseline scores to identify types and direction of causal relations between a pair of biomedical concepts in clinical notes; communicated implicitly or explicitly, identified either in a single sentence or across multiple sentences. We annotate a total of 2714 de-identified examples sampled from the 2018 n2c2 shared task dataset and train four different language model based architectures. Annotation based on our guidelines achieved a high inter-annotator agreement i.e. Fleiss' kappa score of 0.72 and our model for identification of causal relation achieved a macro F1 score of 0.56 on test data. The high inter-annotator agreement for clinical text shows the quality of our annotation guidelines while the provided baseline F1 score sets the direction for future research towards understanding narratives in clinical texts.
Generating Interpretable Counterfactual Explanations By Implicit Minimisation of Epistemic and Aleatoric Uncertainties
Mar 16, 2021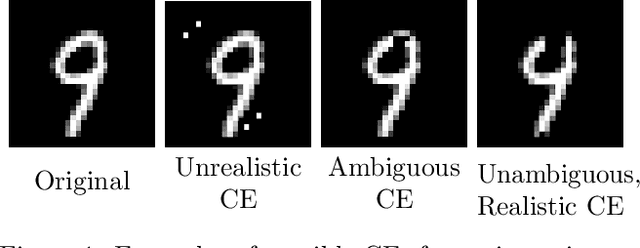
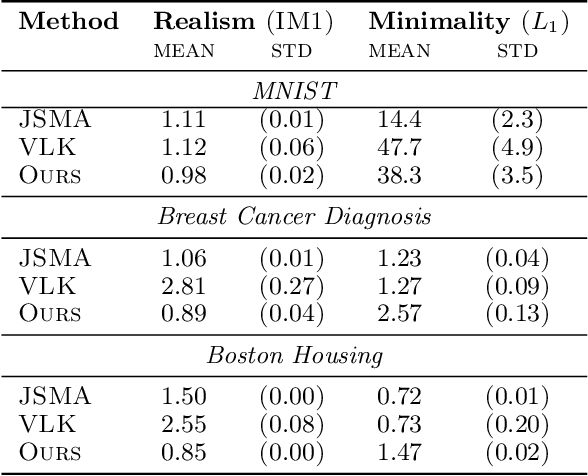

Abstract:Counterfactual explanations (CEs) are a practical tool for demonstrating why machine learning classifiers make particular decisions. For CEs to be useful, it is important that they are easy for users to interpret. Existing methods for generating interpretable CEs rely on auxiliary generative models, which may not be suitable for complex datasets, and incur engineering overhead. We introduce a simple and fast method for generating interpretable CEs in a white-box setting without an auxiliary model, by using the predictive uncertainty of the classifier. Our experiments show that our proposed algorithm generates more interpretable CEs, according to IM1 scores, than existing methods. Additionally, our approach allows us to estimate the uncertainty of a CE, which may be important in safety-critical applications, such as those in the medical domain.
* 21 pages, 13 Figures
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge