Bernardo Bizzo
Deep Metric Learning-based Image Retrieval System for Chest Radiograph and its Clinical Applications in COVID-19
Nov 26, 2020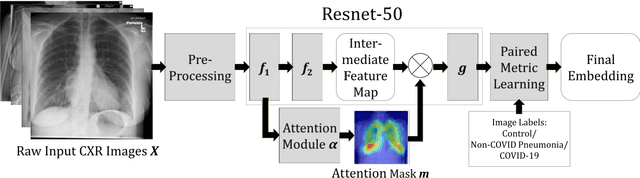
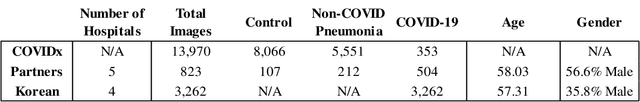
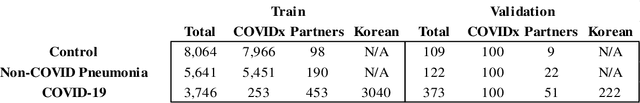
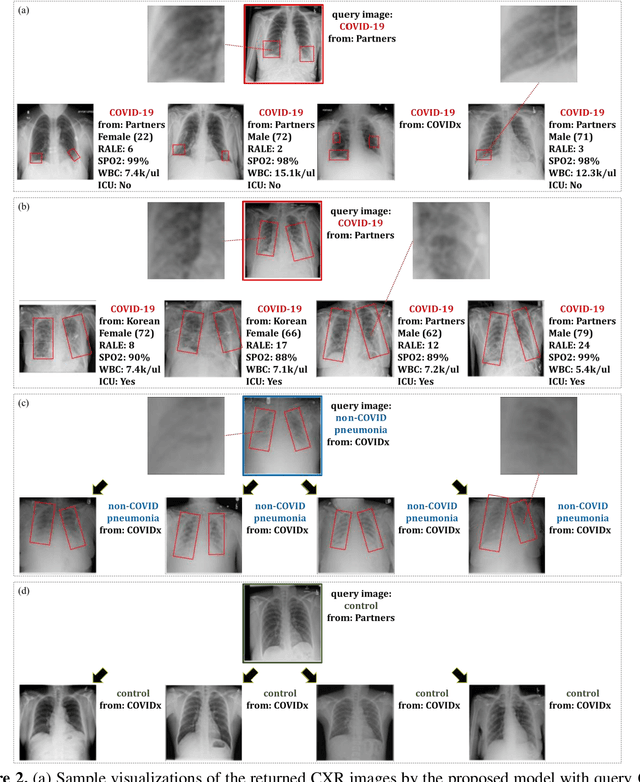
Abstract:In recent years, deep learning-based image analysis methods have been widely applied in computer-aided detection, diagnosis and prognosis, and has shown its value during the public health crisis of the novel coronavirus disease 2019 (COVID-19) pandemic. Chest radiograph (CXR) has been playing a crucial role in COVID-19 patient triaging, diagnosing and monitoring, particularly in the United States. Considering the mixed and unspecific signals in CXR, an image retrieval model of CXR that provides both similar images and associated clinical information can be more clinically meaningful than a direct image diagnostic model. In this work we develop a novel CXR image retrieval model based on deep metric learning. Unlike traditional diagnostic models which aims at learning the direct mapping from images to labels, the proposed model aims at learning the optimized embedding space of images, where images with the same labels and similar contents are pulled together. It utilizes multi-similarity loss with hard-mining sampling strategy and attention mechanism to learn the optimized embedding space, and provides similar images to the query image. The model is trained and validated on an international multi-site COVID-19 dataset collected from 3 different sources. Experimental results of COVID-19 image retrieval and diagnosis tasks show that the proposed model can serve as a robust solution for CXR analysis and patient management for COVID-19. The model is also tested on its transferability on a different clinical decision support task, where the pre-trained model is applied to extract image features from a new dataset without any further training. These results demonstrate our deep metric learning based image retrieval model is highly efficient in the CXR retrieval, diagnosis and prognosis, and thus has great clinical value for the treatment and management of COVID-19 patients.
Semi-Supervised Natural Language Approach for Fine-Grained Classification of Medical Reports
Nov 14, 2019
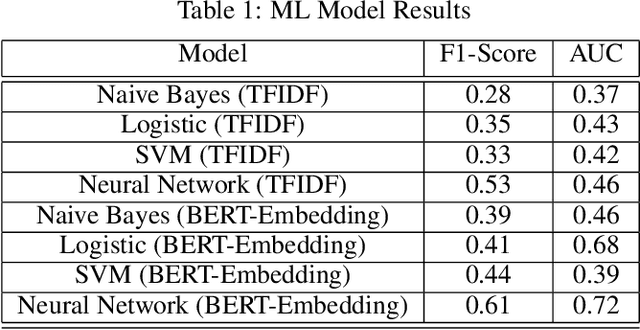
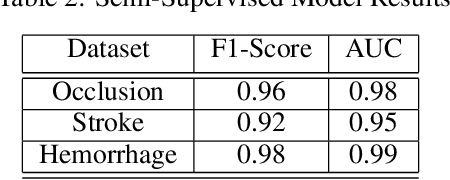
Abstract:Although machine learning has become a powerful tool to augment doctors in clinical analysis, the immense amount of labeled data that is necessary to train supervised learning approaches burdens each development task as time and resource intensive. The vast majority of dense clinical information is stored in written reports, detailing pertinent patient information. The challenge with utilizing natural language data for standard model development is due to the complex nature of the modality. In this research, a model pipeline was developed to utilize an unsupervised approach to train an encoder-language model, a recurrent network, to generate document encodings; which then can be used as features passed into a decoder-classifier model that requires magnitudes less labeled data than previous approaches to differentiate between fine-grained disease classes accurately. The language model was trained on unlabeled radiology reports from the Massachusetts General Hospital Radiology Department (n=218,159) and terminated with a loss of 1.62. The classification models were trained on three labeled datasets of head CT studies of reported patients, presenting large vessel occlusion (n=1403), acute ischemic strokes (n=331), and intracranial hemorrhage (n=4350), to identify a variety of different findings directly from the radiology report data; resulting in AUCs of 0.98, 0.95, and 0.99, respectively, for the large vessel occlusion, acute ischemic stroke, and intracranial hemorrhage datasets. The output encodings are able to be used in conjunction with imaging data, to create models that can process a multitude of different modalities. The ability to automatically extract relevant features from textual data allows for faster model development and integration of textual modality, overall, allowing clinical reports to become a more viable input for more encompassing and accurate deep learning models.
DeepSPINE: Automated Lumbar Vertebral Segmentation, Disc-level Designation, and Spinal Stenosis Grading Using Deep Learning
Jul 26, 2018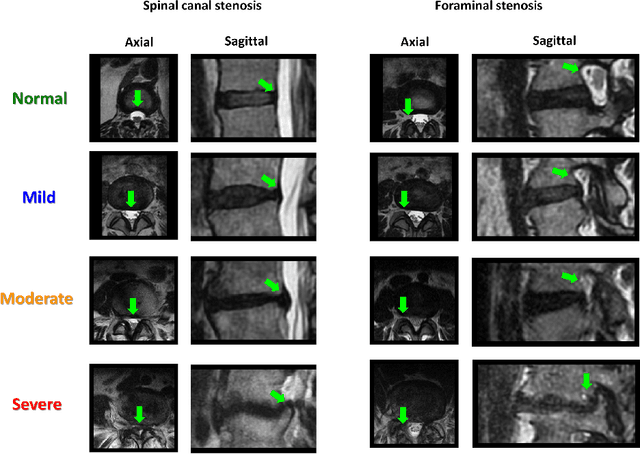
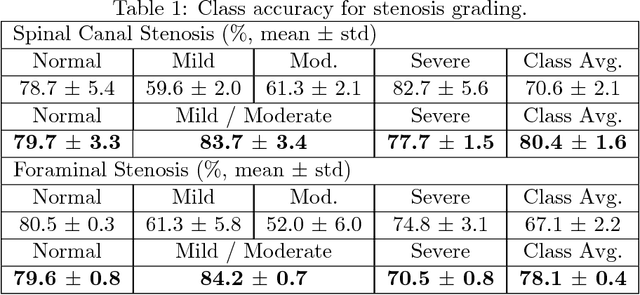
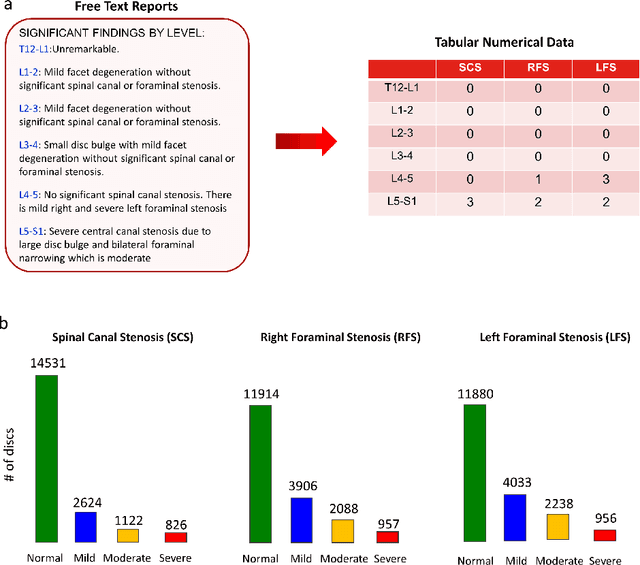
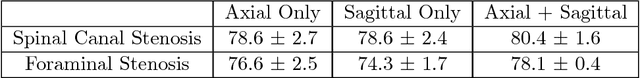
Abstract:The high prevalence of spinal stenosis results in a large volume of MRI imaging, yet interpretation can be time-consuming with high inter-reader variability even among the most specialized radiologists. In this paper, we develop an efficient methodology to leverage the subject-matter-expertise stored in large-scale archival reporting and image data for a deep-learning approach to fully-automated lumbar spinal stenosis grading. Specifically, we introduce three major contributions: (1) a natural-language-processing scheme to extract level-by-level ground-truth labels from free-text radiology reports for the various types and grades of spinal stenosis (2) accurate vertebral segmentation and disc-level localization using a U-Net architecture combined with a spine-curve fitting method, and (3) a multi-input, multi-task, and multi-class convolutional neural network to perform central canal and foraminal stenosis grading on both axial and sagittal imaging series inputs with the extracted report-derived labels applied to corresponding imaging level segments. This study uses a large dataset of 22796 disc-levels extracted from 4075 patients. We achieve state-of-the-art performance on lumbar spinal stenosis classification and expect the technique will increase both radiology workflow efficiency and the perceived value of radiology reports for referring clinicians and patients.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge