Romane Gauriau
Phenotype Detection in Real World Data via Online MixEHR Algorithm
Nov 15, 2022



Abstract:Understanding patterns of diagnoses, medications, procedures, and laboratory tests from electronic health records (EHRs) and health insurer claims is important for understanding disease risk and for efficient clinical development, which often require rules-based curation in collaboration with clinicians. We extended an unsupervised phenotyping algorithm, mixEHR, to an online version allowing us to use it on order of magnitude larger datasets including a large, US-based claims dataset and a rich regional EHR dataset. In addition to recapitulating previously observed disease groups, we discovered clinically meaningful disease subtypes and comorbidities. This work scaled up an effective unsupervised learning method, reinforced existing clinical knowledge, and is a promising approach for efficient collaboration with clinicians.
"Name that manufacturer". Relating image acquisition bias with task complexity when training deep learning models: experiments on head CT
Aug 19, 2020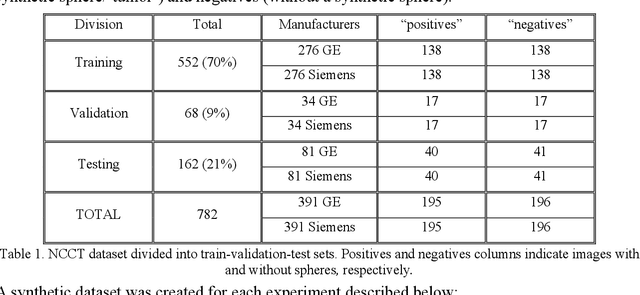
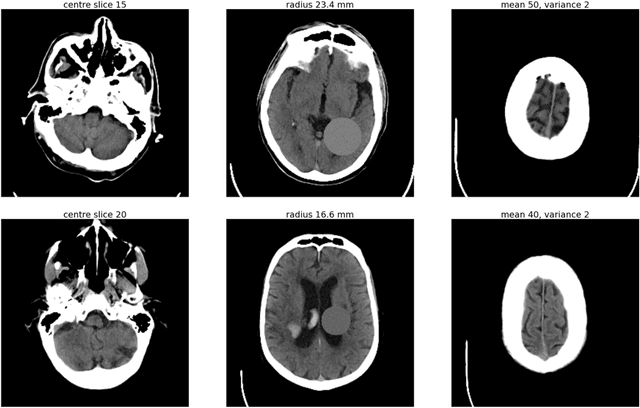
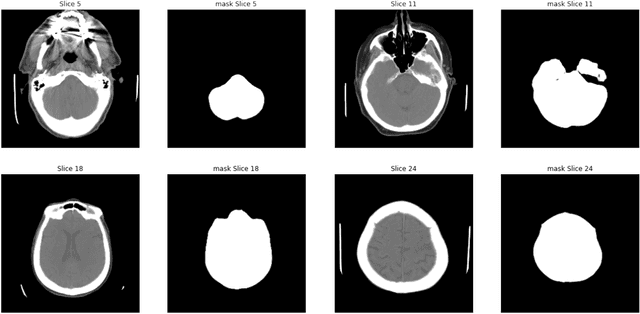

Abstract:As interest in applying machine learning techniques for medical images continues to grow at a rapid pace, models are starting to be developed and deployed for clinical applications. In the clinical AI model development lifecycle (described by Lu et al. [1]), a crucial phase for machine learning scientists and clinicians is the proper design and collection of the data cohort. The ability to recognize various forms of biases and distribution shifts in the dataset is critical at this step. While it remains difficult to account for all potential sources of bias, techniques can be developed to identify specific types of bias in order to mitigate their impact. In this work we analyze how the distribution of scanner manufacturers in a dataset can contribute to the overall bias of deep learning models. We evaluate convolutional neural networks (CNN) for both classification and segmentation tasks, specifically two state-of-the-art models: ResNet [2] for classification and U-Net [3] for segmentation. We demonstrate that CNNs can learn to distinguish the imaging scanner manufacturer and that this bias can substantially impact model performance for both classification and segmentation tasks. By creating an original synthesis dataset of brain data mimicking the presence of more or less subtle lesions we also show that this bias is related to the difficulty of the task. Recognition of such bias is critical to develop robust, generalizable models that will be crucial for clinical applications in real-world data distributions.
An Overview and Case Study of the Clinical AI Model Development Life Cycle for Healthcare Systems
Mar 26, 2020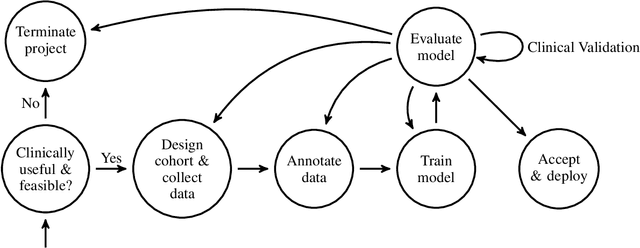
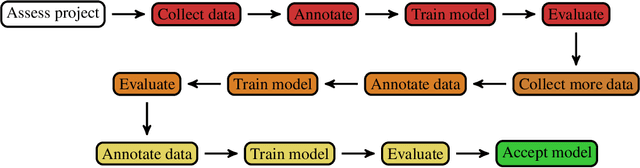
Abstract:Healthcare is one of the most promising areas for machine learning models to make a positive impact. However, successful adoption of AI-based systems in healthcare depends on engaging and educating stakeholders from diverse backgrounds about the development process of AI models. We present a broadly accessible overview of the development life cycle of clinical AI models that is general enough to be adapted to most machine learning projects, and then give an in-depth case study of the development process of a deep learning based system to detect aortic aneurysms in Computed Tomography (CT) exams. We hope other healthcare institutions and clinical practitioners find the insights we share about the development process useful in informing their own model development efforts and to increase the likelihood of successful deployment and integration of AI in healthcare.
Semi-Supervised Natural Language Approach for Fine-Grained Classification of Medical Reports
Nov 14, 2019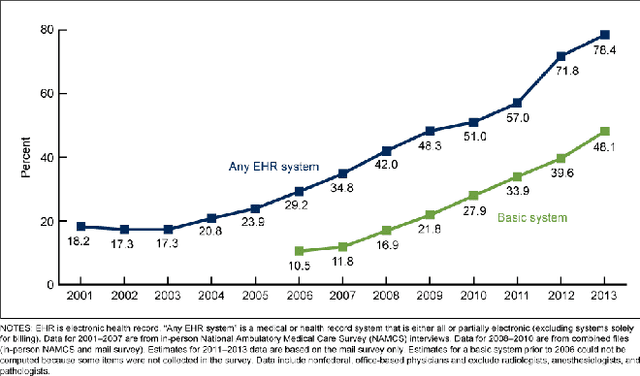
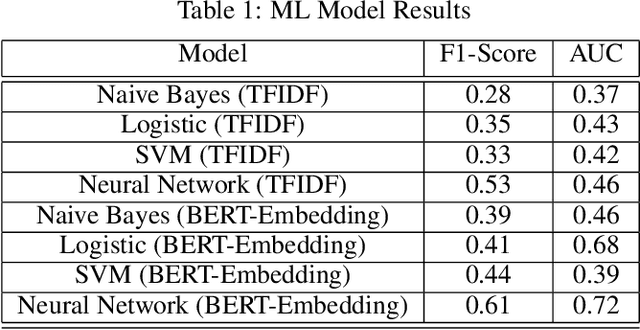
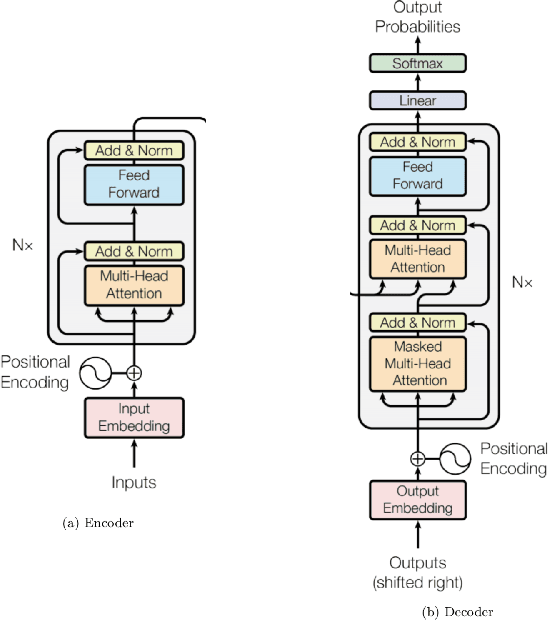
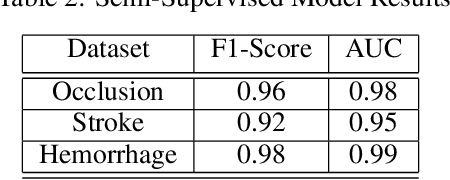
Abstract:Although machine learning has become a powerful tool to augment doctors in clinical analysis, the immense amount of labeled data that is necessary to train supervised learning approaches burdens each development task as time and resource intensive. The vast majority of dense clinical information is stored in written reports, detailing pertinent patient information. The challenge with utilizing natural language data for standard model development is due to the complex nature of the modality. In this research, a model pipeline was developed to utilize an unsupervised approach to train an encoder-language model, a recurrent network, to generate document encodings; which then can be used as features passed into a decoder-classifier model that requires magnitudes less labeled data than previous approaches to differentiate between fine-grained disease classes accurately. The language model was trained on unlabeled radiology reports from the Massachusetts General Hospital Radiology Department (n=218,159) and terminated with a loss of 1.62. The classification models were trained on three labeled datasets of head CT studies of reported patients, presenting large vessel occlusion (n=1403), acute ischemic strokes (n=331), and intracranial hemorrhage (n=4350), to identify a variety of different findings directly from the radiology report data; resulting in AUCs of 0.98, 0.95, and 0.99, respectively, for the large vessel occlusion, acute ischemic stroke, and intracranial hemorrhage datasets. The output encodings are able to be used in conjunction with imaging data, to create models that can process a multitude of different modalities. The ability to automatically extract relevant features from textual data allows for faster model development and integration of textual modality, overall, allowing clinical reports to become a more viable input for more encompassing and accurate deep learning models.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge