Anna Decker
Phenotype Detection in Real World Data via Online MixEHR Algorithm
Nov 15, 2022



Abstract:Understanding patterns of diagnoses, medications, procedures, and laboratory tests from electronic health records (EHRs) and health insurer claims is important for understanding disease risk and for efficient clinical development, which often require rules-based curation in collaboration with clinicians. We extended an unsupervised phenotyping algorithm, mixEHR, to an online version allowing us to use it on order of magnitude larger datasets including a large, US-based claims dataset and a rich regional EHR dataset. In addition to recapitulating previously observed disease groups, we discovered clinically meaningful disease subtypes and comorbidities. This work scaled up an effective unsupervised learning method, reinforced existing clinical knowledge, and is a promising approach for efficient collaboration with clinicians.
Identification of Latent Variables From Graphical Model Residuals
Jan 07, 2021
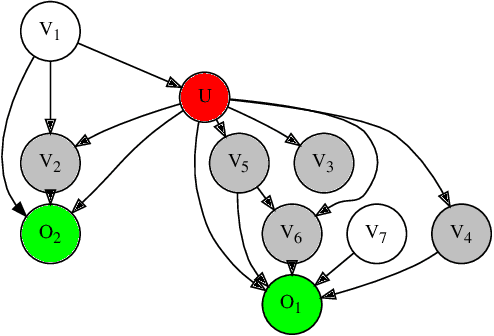
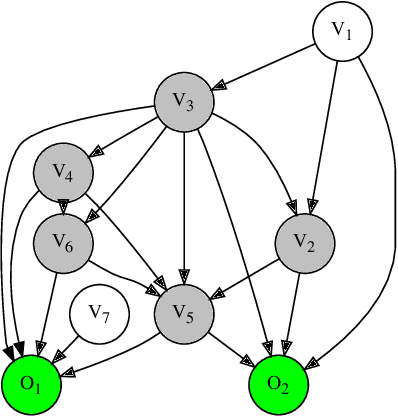
Abstract:Graph-based causal discovery methods aim to capture conditional independencies consistent with the observed data and differentiate causal relationships from indirect or induced ones. Successful construction of graphical models of data depends on the assumption of causal sufficiency: that is, that all confounding variables are measured. When this assumption is not met, learned graphical structures may become arbitrarily incorrect and effects implied by such models may be wrongly attributed, carry the wrong magnitude, or mis-represent direction of correlation. Wide application of graphical models to increasingly less curated "big data" draws renewed attention to the unobserved confounder problem. We present a novel method that aims to control for the latent space when estimating a DAG by iteratively deriving proxies for the latent space from the residuals of the inferred model. Under mild assumptions, our method improves structural inference of Gaussian graphical models and enhances identifiability of the causal effect. In addition, when the model is being used to predict outcomes, it un-confounds the coefficients on the parents of the outcomes and leads to improved predictive performance when out-of-sample regime is very different from the training data. We show that any improvement of prediction of an outcome is intrinsically capped and cannot rise beyond a certain limit as compared to the confounded model. We extend our methodology beyond GGMs to ordinal variables and nonlinear cases. Our R package provides both PCA and autoencoder implementations of the methodology, suitable for GGMs with some guarantees and for better performance in general cases but without such guarantees.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge