Atanas Mirchev
Filter-Aware Model-Predictive Control
Apr 20, 2023Abstract:Partially-observable problems pose a trade-off between reducing costs and gathering information. They can be solved optimally by planning in belief space, but that is often prohibitively expensive. Model-predictive control (MPC) takes the alternative approach of using a state estimator to form a belief over the state, and then plan in state space. This ignores potential future observations during planning and, as a result, cannot actively increase or preserve the certainty of its own state estimate. We find a middle-ground between planning in belief space and completely ignoring its dynamics by only reasoning about its future accuracy. Our approach, filter-aware MPC, penalises the loss of information by what we call "trackability", the expected error of the state estimator. We show that model-based simulation allows condensing trackability into a neural network, which allows fast planning. In experiments involving visual navigation, realistic every-day environments and a two-link robot arm, we show that filter-aware MPC vastly improves regular MPC.
PRISM: Probabilistic Real-Time Inference in Spatial World Models
Dec 06, 2022Abstract:We introduce PRISM, a method for real-time filtering in a probabilistic generative model of agent motion and visual perception. Previous approaches either lack uncertainty estimates for the map and agent state, do not run in real-time, do not have a dense scene representation or do not model agent dynamics. Our solution reconciles all of these aspects. We start from a predefined state-space model which combines differentiable rendering and 6-DoF dynamics. Probabilistic inference in this model amounts to simultaneous localisation and mapping (SLAM) and is intractable. We use a series of approximations to Bayesian inference to arrive at probabilistic map and state estimates. We take advantage of well-established methods and closed-form updates, preserving accuracy and enabling real-time capability. The proposed solution runs at 10Hz real-time and is similarly accurate to state-of-the-art SLAM in small to medium-sized indoor environments, with high-speed UAV and handheld camera agents (Blackbird, EuRoC and TUM-RGBD).
Tracking and Planning with Spatial World Models
Jan 25, 2022



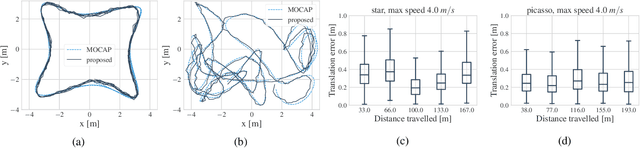
Abstract:We introduce a method for real-time navigation and tracking with differentiably rendered world models. Learning models for control has led to impressive results in robotics and computer games, but this success has yet to be extended to vision-based navigation. To address this, we transfer advances in the emergent field of differentiable rendering to model-based control. We do this by planning in a learned 3D spatial world model, combined with a pose estimation algorithm previously used in the context of TSDF fusion, but now tailored to our setting and improved to incorporate agent dynamics. We evaluate over six simulated environments based on complex human-designed floor plans and provide quantitative results. We achieve up to 92% navigation success rate at a frequency of 15 Hz using only image and depth observations under stochastic, continuous dynamics.
Mind the Gap when Conditioning Amortised Inference in Sequential Latent-Variable Models
Jan 18, 2021



Abstract:Amortised inference enables scalable learning of sequential latent-variable models (LVMs) with the evidence lower bound (ELBO). In this setting, variational posteriors are often only partially conditioned. While the true posteriors depend, e.g., on the entire sequence of observations, approximate posteriors are only informed by past observations. This mimics the Bayesian filter -- a mixture of smoothing posteriors. Yet, we show that the ELBO objective forces partially-conditioned amortised posteriors to approximate products of smoothing posteriors instead. Consequently, the learned generative model is compromised. We demonstrate these theoretical findings in three scenarios: traffic flow, handwritten digits, and aerial vehicle dynamics. Using fully-conditioned approximate posteriors, performance improves in terms of generative modelling and multi-step prediction.
Variational State-Space Models for Localisation and Dense 3D Mapping in 6 DoF
Jun 17, 2020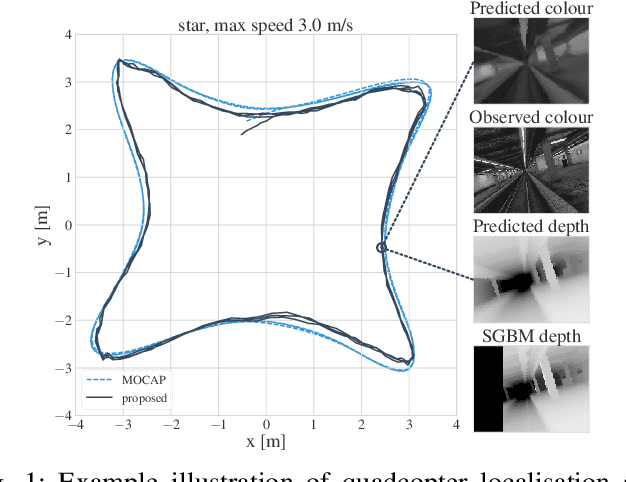
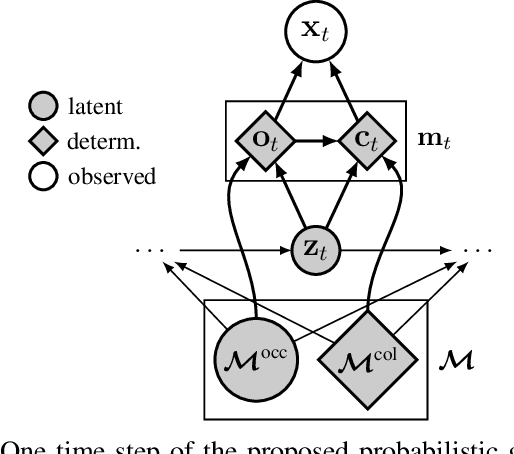
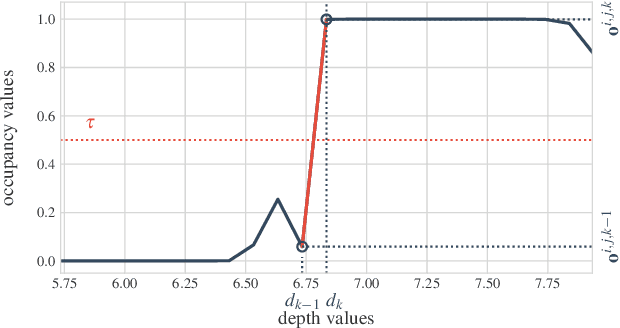
Abstract:We solve the problem of 6-DoF localisation and 3D dense reconstruction in spatial environments as approximate Bayesian inference in a deep generative approach which combines learned with engineered models. This principled treatment of uncertainty and probabilistic inference overcomes the shortcoming of current state-of-the-art solutions to rely on heavily engineered, heterogeneous pipelines. Variational inference enables us to use neural networks for system identification, while a differentiable raycaster is used for the emission model. This ensures that our model is amenable to end-to-end gradient-based optimisation. We evaluate our approach on realistic unmanned aerial vehicle flight data, nearing the performance of a state-of-the-art visual inertial odometry system. The applicability of the learned model to downstream tasks such as generative prediction and planning is investigated.
Approximate Bayesian inference in spatial environments
May 18, 2018



Abstract:We propose to learn a stochastic recurrent model to solve the problem of simultaneous localisation and mapping (SLAM). Our model is a deep variational Bayes filter augmented with a latent global variable---similar to an external memory component---representing the spatially structured environment. Reasoning about the pose of an agent and the map of the environment is then naturally expressed as posterior inference in the resulting generative model. We evaluate the method on a set of randomly generated mazes which are traversed by an agent equipped with laser range finders. Path integration based on an accurate motion model is consistently outperformed, and most importantly, drift practically eliminated. Our approach inherits favourable properties from neural networks, such as differentiability, flexibility and the ability to train components either in isolation or end-to-end.
Classification of sparsely labeled spatio-temporal data through semi-supervised adversarial learning
Jan 29, 2018



Abstract:In recent years, Generative Adversarial Networks (GAN) have emerged as a powerful method for learning the mapping from noisy latent spaces to realistic data samples in high-dimensional space. So far, the development and application of GANs have been predominantly focused on spatial data such as images. In this project, we aim at modeling of spatio-temporal sensor data instead, i.e. dynamic data over time. The main goal is to encode temporal data into a global and low-dimensional latent vector that captures the dynamics of the spatio-temporal signal. To this end, we incorporate auto-regressive RNNs, Wasserstein GAN loss, spectral norm weight constraints and a semi-supervised learning scheme into InfoGAN, a method for retrieval of meaningful latents in adversarial learning. To demonstrate the modeling capability of our method, we encode full-body skeletal human motion from a large dataset representing 60 classes of daily activities, recorded in a multi-Kinect setup. Initial results indicate competitive classification performance of the learned latent representations, compared to direct CNN/RNN inference. In future work, we plan to apply this method on a related problem in the medical domain, i.e. on recovery of meaningful latents in gait analysis of patients with vertigo and balance disorders.
3D Deep Learning for Biological Function Prediction from Physical Fields
Apr 13, 2017
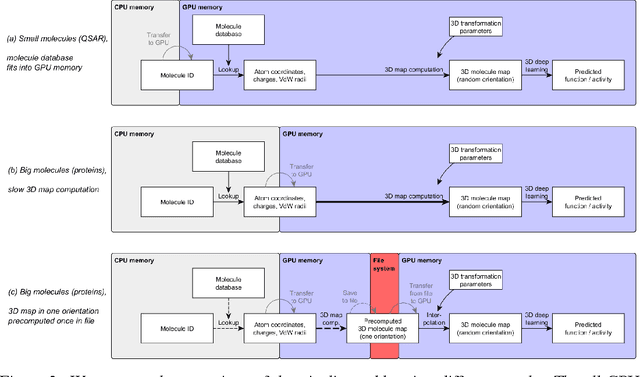
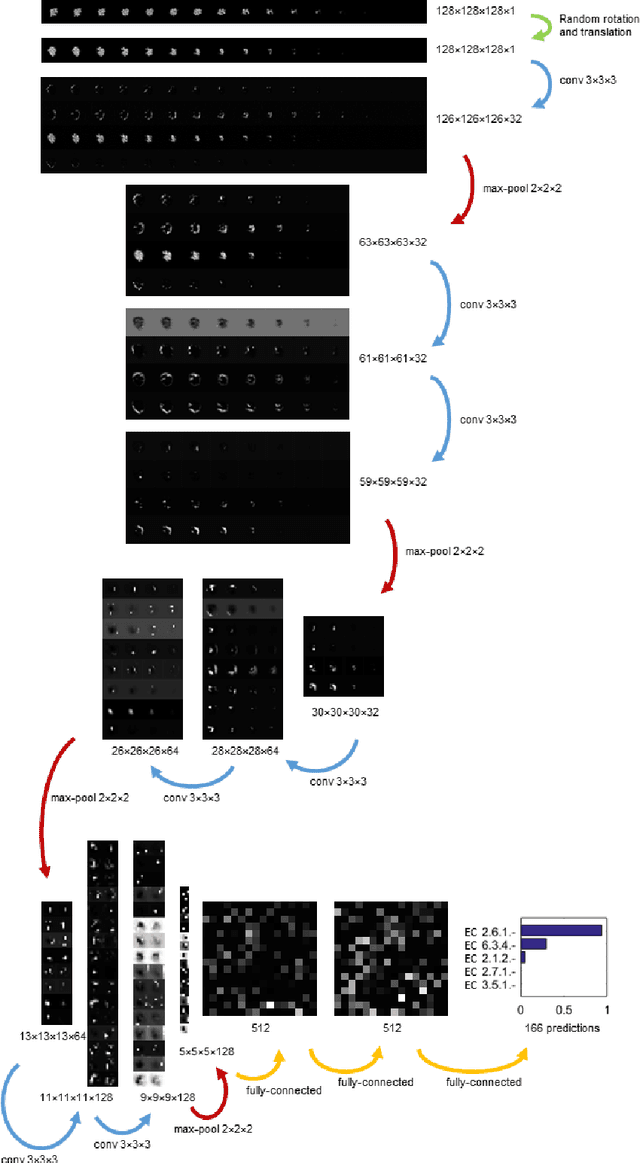
Abstract:Predicting the biological function of molecules, be it proteins or drug-like compounds, from their atomic structure is an important and long-standing problem. Function is dictated by structure, since it is by spatial interactions that molecules interact with each other, both in terms of steric complementarity, as well as intermolecular forces. Thus, the electron density field and electrostatic potential field of a molecule contain the "raw fingerprint" of how this molecule can fit to binding partners. In this paper, we show that deep learning can predict biological function of molecules directly from their raw 3D approximated electron density and electrostatic potential fields. Protein function based on EC numbers is predicted from the approximated electron density field. In another experiment, the activity of small molecules is predicted with quality comparable to state-of-the-art descriptor-based methods. We propose several alternative computational models for the GPU with different memory and runtime requirements for different sizes of molecules and of databases. We also propose application-specific multi-channel data representations. With future improvements of training datasets and neural network settings in combination with complementary information sources (sequence, genomic context, expression level), deep learning can be expected to show its generalization power and revolutionize the field of molecular function prediction.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge