Georgi Dikov
FastCAD: Real-Time CAD Retrieval and Alignment from Scans and Videos
Mar 22, 2024



Abstract:Digitising the 3D world into a clean, CAD model-based representation has important applications for augmented reality and robotics. Current state-of-the-art methods are computationally intensive as they individually encode each detected object and optimise CAD alignments in a second stage. In this work, we propose FastCAD, a real-time method that simultaneously retrieves and aligns CAD models for all objects in a given scene. In contrast to previous works, we directly predict alignment parameters and shape embeddings. We achieve high-quality shape retrievals by learning CAD embeddings in a contrastive learning framework and distilling those into FastCAD. Our single-stage method accelerates the inference time by a factor of 50 compared to other methods operating on RGB-D scans while outperforming them on the challenging Scan2CAD alignment benchmark. Further, our approach collaborates seamlessly with online 3D reconstruction techniques. This enables the real-time generation of precise CAD model-based reconstructions from videos at 10 FPS. Doing so, we significantly improve the Scan2CAD alignment accuracy in the video setting from 43.0% to 48.2% and the reconstruction accuracy from 22.9% to 29.6%.
Calibrated Adversarial Refinement for Multimodal Semantic Segmentation
Jun 23, 2020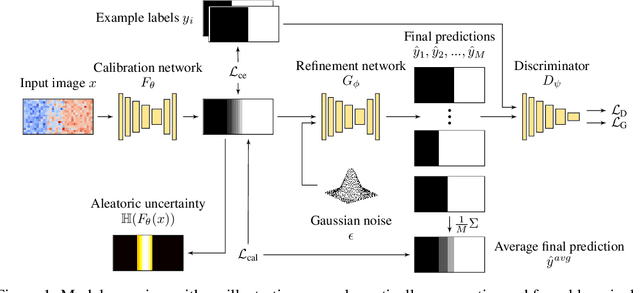
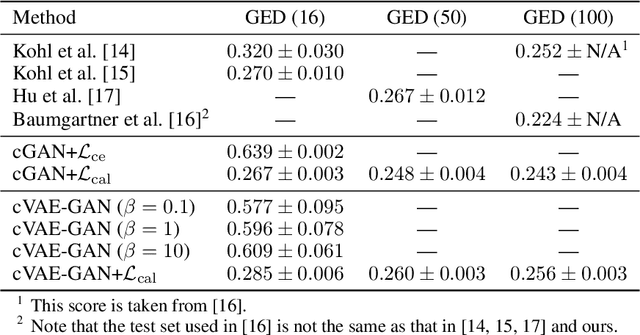
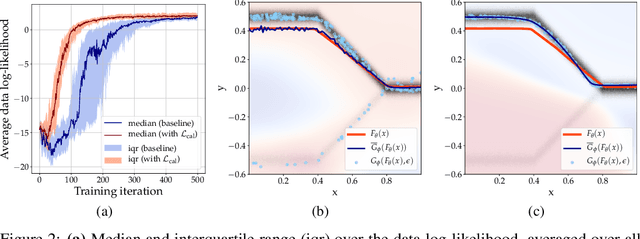
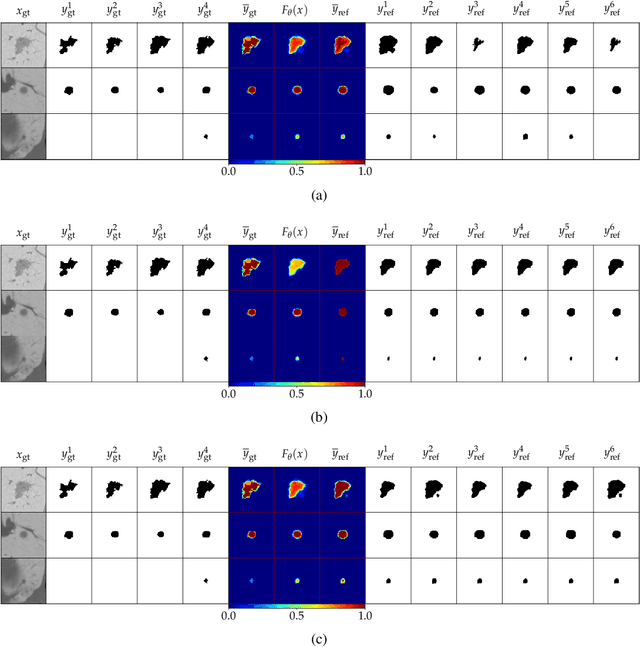
Abstract:Ambiguities in images or unsystematic annotation can lead to multiple valid solutions in semantic segmentation. To learn a distribution over predictions, recent work has explored the use of probabilistic networks. However, these do not necessarily capture the empirical distribution accurately. In this work, we aim to learn a calibrated multimodal predictive distribution, where the empirical frequency of the sampled predictions closely reflects that of the corresponding labels in the training set. To this end, we propose a novel two-stage, cascaded strategy for calibrated adversarial refinement. In the first stage, we explicitly model the data with a categorical likelihood. In the second, we train an adversarial network to sample from it an arbitrary number of coherent predictions. The model can be used independently or integrated into any black-box segmentation framework to enable the synthesis of diverse predictions. We demonstrate the utility and versatility of the approach by achieving competitive results on the multigrader LIDC dataset and a modified Cityscapes dataset. In addition, we use a toy regression dataset to show that our framework is not confined to semantic segmentation, and the core design can be adapted to other tasks requiring learning a calibrated predictive distribution.
Bayesian Learning of Neural Network Architectures
Jan 27, 2019



Abstract:In this paper we propose a Bayesian method for estimating architectural parameters of neural networks, namely layer size and network depth. We do this by learning concrete distributions over these parameters. Our results show that regular networks with a learnt structure can generalise better on small datasets, while fully stochastic networks can be more robust to parameter initialisation. The proposed method relies on standard neural variational learning and, unlike randomised architecture search, does not require a retraining of the model, thus keeping the computational overhead at minimum.
3D Deep Learning for Biological Function Prediction from Physical Fields
Apr 13, 2017
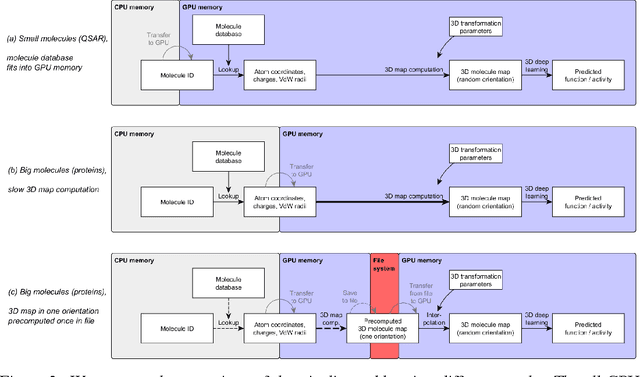
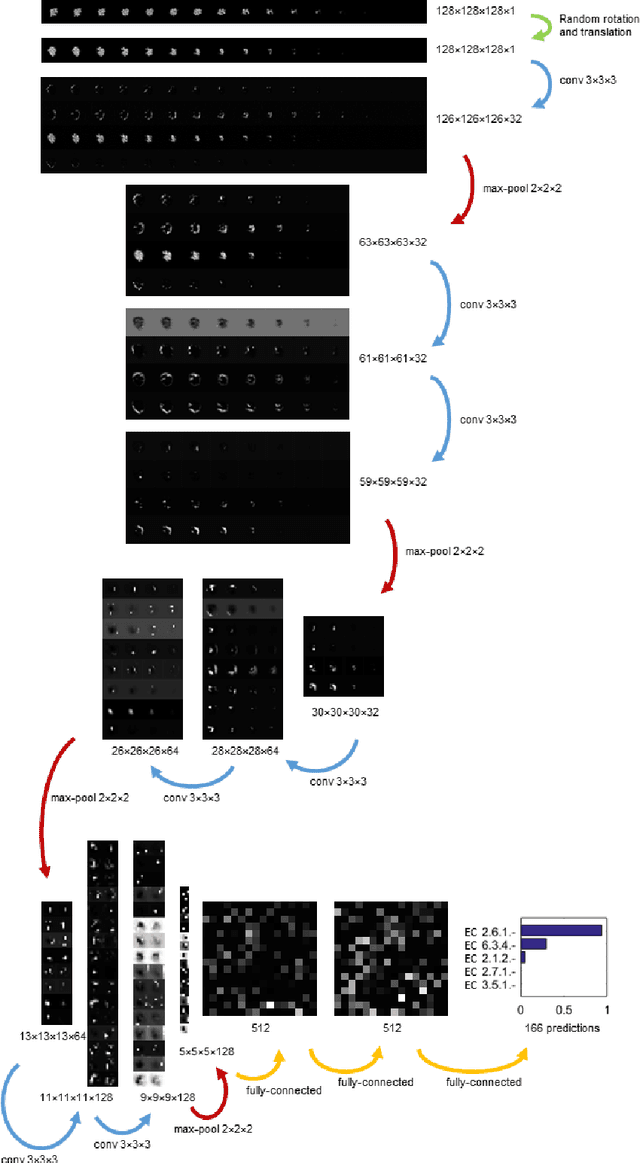
Abstract:Predicting the biological function of molecules, be it proteins or drug-like compounds, from their atomic structure is an important and long-standing problem. Function is dictated by structure, since it is by spatial interactions that molecules interact with each other, both in terms of steric complementarity, as well as intermolecular forces. Thus, the electron density field and electrostatic potential field of a molecule contain the "raw fingerprint" of how this molecule can fit to binding partners. In this paper, we show that deep learning can predict biological function of molecules directly from their raw 3D approximated electron density and electrostatic potential fields. Protein function based on EC numbers is predicted from the approximated electron density field. In another experiment, the activity of small molecules is predicted with quality comparable to state-of-the-art descriptor-based methods. We propose several alternative computational models for the GPU with different memory and runtime requirements for different sizes of molecules and of databases. We also propose application-specific multi-channel data representations. With future improvements of training datasets and neural network settings in combination with complementary information sources (sequence, genomic context, expression level), deep learning can be expected to show its generalization power and revolutionize the field of molecular function prediction.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge